

You can:
| Name | Kappa-type opioid receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | OPRK1 |
| Synonym | K-OR-1 KOPr OP2 KOP KOR-1 [ Show all ] |
| Disease | Obesity Opiate dependence Inflammatory bowel disease Erythema Diarrhea-predominant IBS [ Show all ] |
| Length | 380 |
| Amino acid sequence | MDSPIQIFRGEPGPTCAPSACLPPNSSAWFPGWAEPDSNGSAGSEDAQLEPAHISPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVISIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFPDDDYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGSREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSTSHSTAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFPLKMRMERQSTSRVRNTVQDPAYLRDIDGMNKPV |
| UniProt | P41145 |
| Protein Data Bank | 6b73, 4djh |
| GPCR-HGmod model | P41145 |
| 3D structure model | This structure is from PDB ID 6b73. |
| BioLiP | BL0402244,BL0402246, BL0224693,BL0224694, BL0402243,BL0402245 |
| Therapeutic Target Database | T60693 |
| ChEMBL | CHEMBL237 |
| IUPHAR | 318 |
| DrugBank | BE0000632 |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 136 | 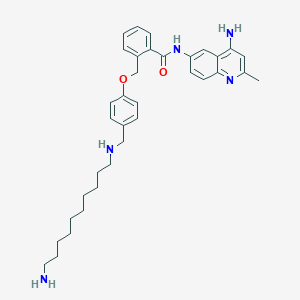 CHEMBL1762411 CHEMBL1762411 | C35H45N5O2 | 567.778 | 6 / 4 | 6.2 | No |
| 168 | 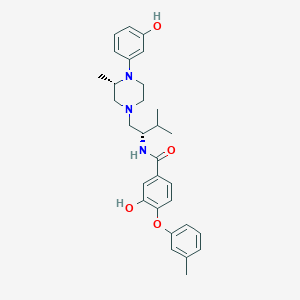 CHEMBL2381177 CHEMBL2381177 | C30H37N3O4 | 503.643 | 6 / 3 | 5.8 | No |
| 441674 |  CHEMBL3409110 CHEMBL3409110 | C29H32FNO2 | 445.578 | 4 / 1 | 7.1 | No |
| 321 | 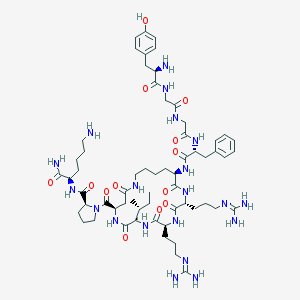 CHEMBL412600 CHEMBL412600 | C61H96N20O13 | 1317.57 | 17 / 18 | -3.5 | No |
| 326 | 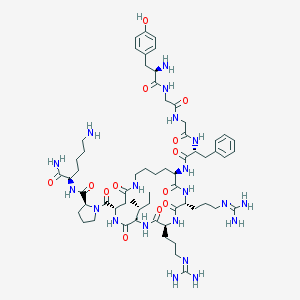 CHEMBL387083 CHEMBL387083 | C61H96N20O13 | 1317.57 | 17 / 18 | -3.5 | No |
| 471 |  CHEMBL1642761 CHEMBL1642761 | C24H30N2 | 346.518 | 2 / 1 | 5.4 | No |
| 568 | 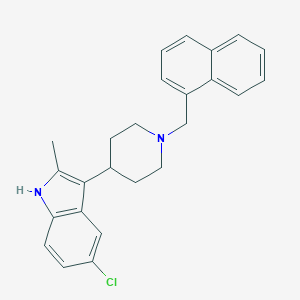 CHEMBL438965 CHEMBL438965 | C25H25ClN2 | 388.939 | 1 / 1 | 6.4 | No |
| 441696 | 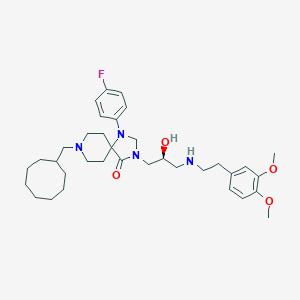 CHEMBL3394753 CHEMBL3394753 | C35H51FN4O4 | 610.815 | 8 / 2 | 6.2 | No |
| 862 | 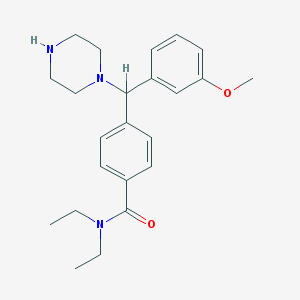 CHEMBL125742 CHEMBL125742 | C23H31N3O2 | 381.52 | 4 / 1 | 3.0 | Yes |
| 947 |  CHEMBL2397017 CHEMBL2397017 | C30H29N3O4 | 495.579 | 6 / 2 | 4.2 | Yes |
| 1014 | 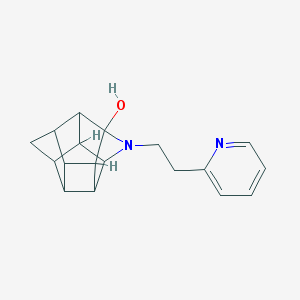 CHEMBL2205816 CHEMBL2205816 | C18H20N2O | 280.371 | 3 / 1 | 1.8 | Yes |
| 1226 | 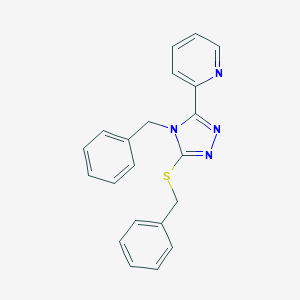 KUC105575N KUC105575N | C21H18N4S | 358.463 | 4 / 0 | 4.0 | Yes |
| 557336 | 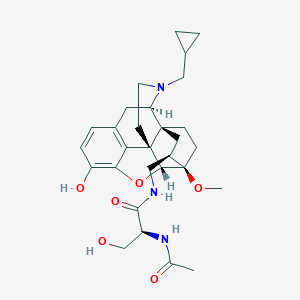 SCHEMBL16034373 SCHEMBL16034373 | C29H39N3O6 | 525.646 | 7 / 4 | 1.7 | No |
| 547915 | 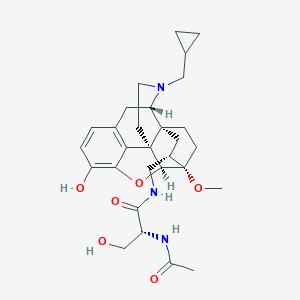 CHEMBL3899312 CHEMBL3899312 | C29H39N3O6 | 525.646 | 7 / 4 | 1.7 | No |
| 557341 |  SCHEMBL10316476 SCHEMBL10316476 | C44H58N8O5 | 778.999 | 7 / 6 | 4.0 | No |
| 547917 | 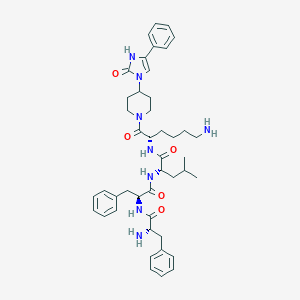 CHEMBL3893631 CHEMBL3893631 | C44H58N8O5 | 778.999 | 7 / 6 | 4.0 | No |
| 1379 | 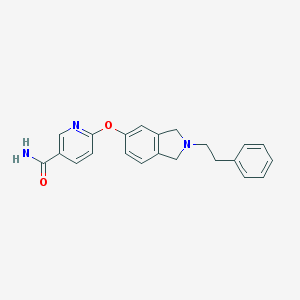 CHEMBL235154 CHEMBL235154 | C22H21N3O2 | 359.429 | 4 / 1 | 3.0 | Yes |
| 535951 | 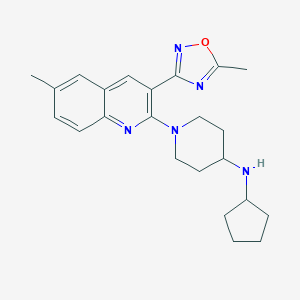 SCHEMBL18993317 SCHEMBL18993317 | C23H29N5O | 391.519 | 6 / 1 | 4.7 | Yes |
| 517326 | 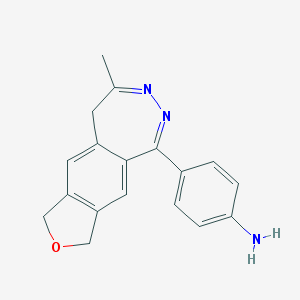 CHEMBL297440 CHEMBL297440 | C18H17N3O | 291.354 | 4 / 1 | 1.7 | Yes |
| 1533 | 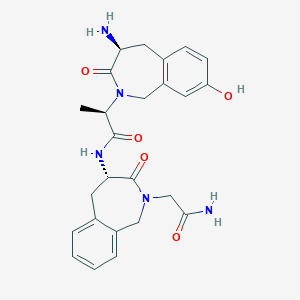 SB-0304 SB-0304 | C25H29N5O5 | 479.537 | 6 / 4 | -0.2 | Yes |
| 521479 | 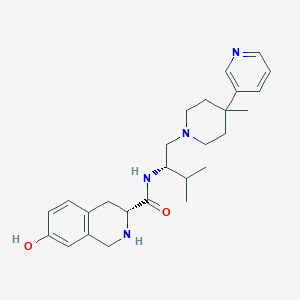 CHEMBL3814950 CHEMBL3814950 | C26H36N4O2 | 436.6 | 5 / 3 | 3.5 | Yes |
| 1566 | 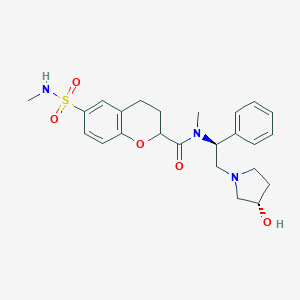 CHEMBL199111 CHEMBL199111 | C24H31N3O5S | 473.588 | 7 / 2 | 1.9 | Yes |
| 547920 | 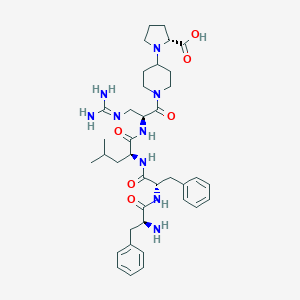 CHEMBL3925949 CHEMBL3925949 | C38H55N9O6 | 733.915 | 9 / 7 | -0.5 | No |
| 557348 | 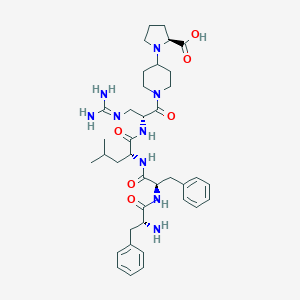 SCHEMBL11932315 SCHEMBL11932315 | C38H55N9O6 | 733.915 | 9 / 7 | -0.5 | No |
| 2082 |  CHEMBL344342 CHEMBL344342 | C19H24Cl2N2O | 367.314 | 2 / 0 | 4.2 | Yes |
| 441758 |  CHEMBL3359280 CHEMBL3359280 | C29H31FO8 | 526.557 | 9 / 0 | 4.3 | No |
| 2149 |  CHEMBL188841 CHEMBL188841 | C14H21NO | 219.328 | 2 / 1 | 3.2 | Yes |
| 521515 |  CHEMBL3759981 CHEMBL3759981 | C43H51N7O7 | 777.923 | 8 / 8 | 3.4 | No |
| 2263 |  1-(cyclohexylmethyl)pyrrolidine 1-(cyclohexylmethyl)pyrrolidine | C11H21N | 167.296 | 1 / 0 | 3.1 | Yes |
| 535981 |  SCHEMBL16944797 SCHEMBL16944797 | C23H29N5O | 391.519 | 6 / 0 | 4.7 | Yes |
| 547923 |  CHEMBL3977305 CHEMBL3977305 | C27H31NO4 | 433.548 | 5 / 2 | 3.8 | Yes |
| 557370 |  SCHEMBL878409 SCHEMBL878409 | C27H31NO4 | 433.548 | 5 / 2 | 3.8 | Yes |
| 547928 |  CHEMBL3945795 CHEMBL3945795 | C15H25NO | 235.371 | 2 / 0 | 2.6 | Yes |
| 2968 |  CHEMBL415223 CHEMBL415223 | C37H44N2O2 | 548.771 | 3 / 2 | 8.1 | No |
| 441841 |  CHEMBL3361419 CHEMBL3361419 | C24H34N6O3 | 454.575 | 6 / 4 | 1.7 | Yes |
| 3181 |  CHEMBL377340 CHEMBL377340 | C18H19N3O | 293.37 | 3 / 1 | 4.1 | Yes |
| 535991 | 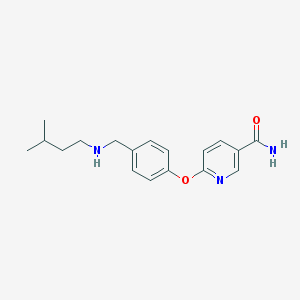 SCHEMBL1532866 SCHEMBL1532866 | C18H23N3O2 | 313.401 | 4 / 2 | 2.6 | Yes |
| 3237 | 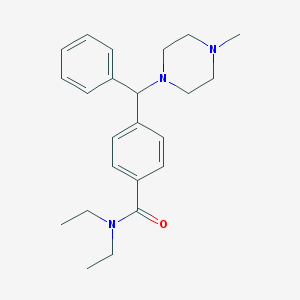 CHEMBL124642 CHEMBL124642 | C23H31N3O | 365.521 | 3 / 0 | 3.5 | Yes |
| 3437 | 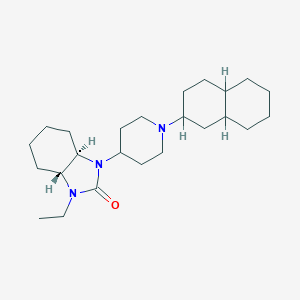 CHEMBL2113273 CHEMBL2113273 | C24H41N3O | 387.612 | 2 / 0 | 5.2 | No |
| 547939 | 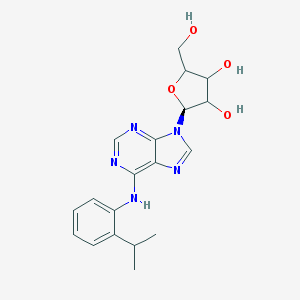 CHEMBL95302 CHEMBL95302 | C19H23N5O4 | 385.424 | 8 / 4 | 2.3 | Yes |
| 3524 | 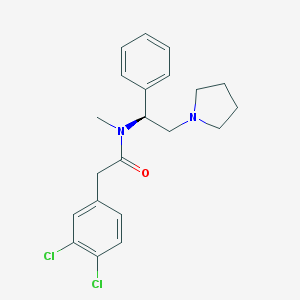 CHEMBL38576 CHEMBL38576 | C21H24Cl2N2O | 391.336 | 2 / 0 | 4.6 | Yes |
| 3533 | 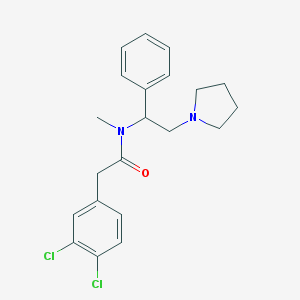 CHEMBL320882 CHEMBL320882 | C21H24Cl2N2O | 391.336 | 2 / 0 | 4.6 | Yes |
| 463319 | 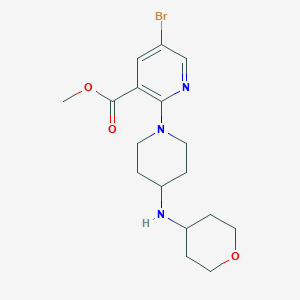 MLS004801940 MLS004801940 | C17H24BrN3O3 | 398.301 | 6 / 1 | 2.4 | Yes |
| 3801 |  CHEMBL127463 CHEMBL127463 | C25H24FN3O | 401.485 | 4 / 1 | 4.7 | Yes |
| 3831 | 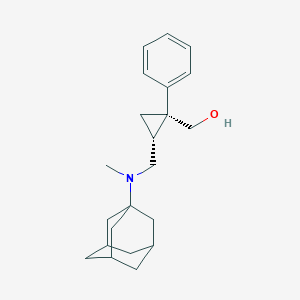 CHEMBL2260849 CHEMBL2260849 | C22H31NO | 325.496 | 2 / 1 | 4.4 | Yes |
| 3866 |  CHEMBL228740 CHEMBL228740 | C22H23NOS | 349.492 | 3 / 1 | 4.3 | Yes |
| 3940 |  CHEMBL364398 CHEMBL364398 | C23H37N3O3S | 435.627 | 6 / 1 | 3.3 | Yes |
| 521557 | 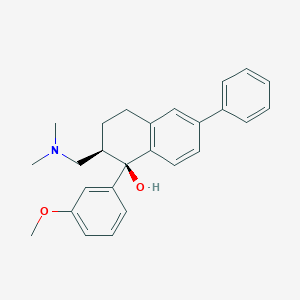 CHEMBL3809808 CHEMBL3809808 | C26H29NO2 | 387.523 | 3 / 1 | 4.8 | Yes |
| 521563 | 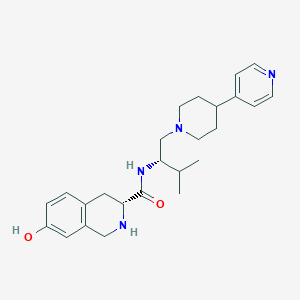 CHEMBL3815013 CHEMBL3815013 | C25H34N4O2 | 422.573 | 5 / 3 | 3.0 | Yes |
| 4485 | 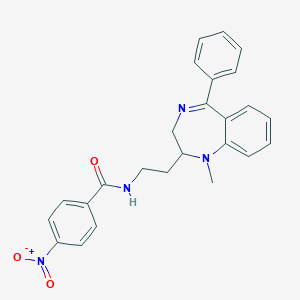 CHEMBL338391 CHEMBL338391 | C25H24N4O3 | 428.492 | 5 / 1 | 4.4 | Yes |
| 536048 | 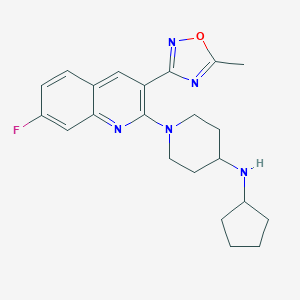 SCHEMBL16945023 SCHEMBL16945023 | C22H26FN5O | 395.482 | 7 / 1 | 4.4 | Yes |
| 4665 | 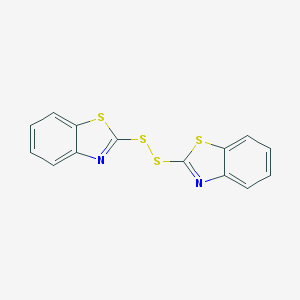 120-78-5 120-78-5 | C14H8N2S4 | 332.472 | 6 / 0 | 5.6 | No |
| 4824 | 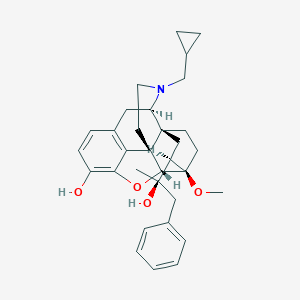 CHEMBL2338752 CHEMBL2338752 | C32H39NO4 | 501.667 | 5 / 2 | 5.2 | No |
| 4922 | 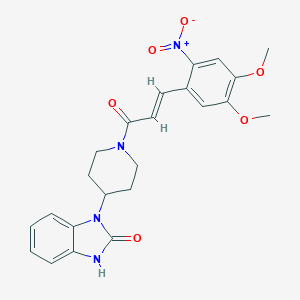 CHEMBL332697 CHEMBL332697 | C23H24N4O6 | 452.467 | 6 / 1 | 2.7 | Yes |
| 4937 |  CHEMBL319589 CHEMBL319589 | C28H29NO4 | 443.543 | 5 / 1 | 5.1 | No |
| 5174 |  MLS000714572 MLS000714572 | C22H30N2O6 | 418.49 | 6 / 2 | 2.5 | Yes |
| 5202 | 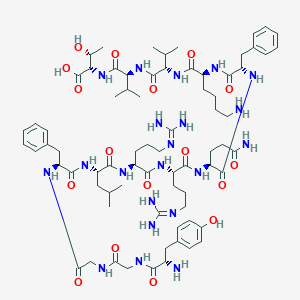 Dynorphin B Dynorphin B | C74H115N21O17 | 1570.86 | 21 / 22 | -3.6 | No |
| 536068 |  CHEMBL3927136 CHEMBL3927136 | C30H42N2O5 | 510.675 | 7 / 2 | 4.4 | No |
| 463526 | 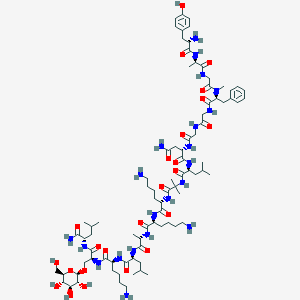 CHEMBL3597077 CHEMBL3597077 | C84H139N21O24 | 1827.16 | 28 / 25 | -4.1 | No |
| 5580 | 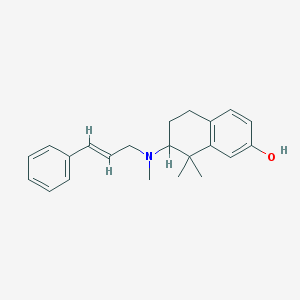 CHEMBL365483 CHEMBL365483 | C22H27NO | 321.464 | 2 / 1 | 5.3 | No |
| 5670 | 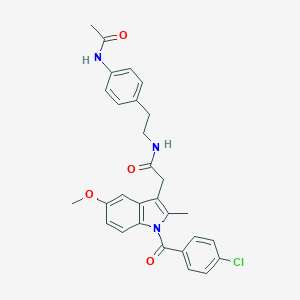 AC1L1HUK AC1L1HUK | C29H28ClN3O4 | 518.01 | 4 / 2 | 5.2 | No |
| 5822 | 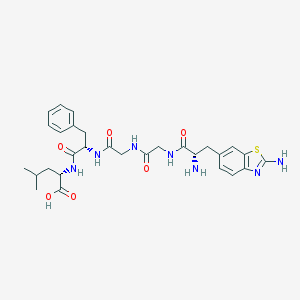 H-Aba-Gly-Gly-Phe-Leu-OH H-Aba-Gly-Gly-Phe-Leu-OH | C29H37N7O6S | 611.718 | 10 / 7 | -0.2 | No |
| 5872 | 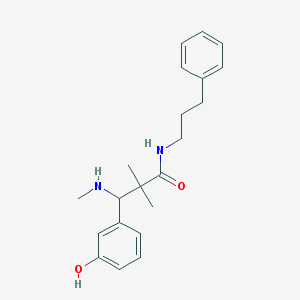 CHEMBL347750 CHEMBL347750 | C21H28N2O2 | 340.467 | 3 / 3 | 3.5 | Yes |
| 557447 | 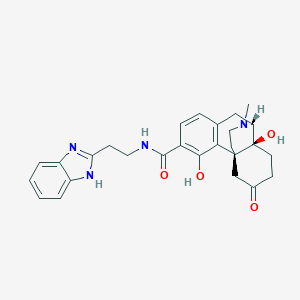 SCHEMBL14466685 SCHEMBL14466685 | C27H30N4O4 | 474.561 | 6 / 4 | 2.3 | Yes |
| 6019 | 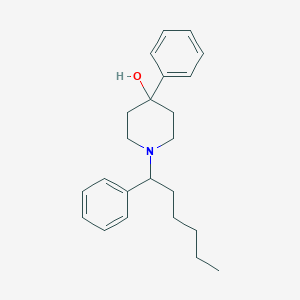 CHEMBL230253 CHEMBL230253 | C23H31NO | 337.507 | 2 / 1 | 5.3 | No |
| 6055 | 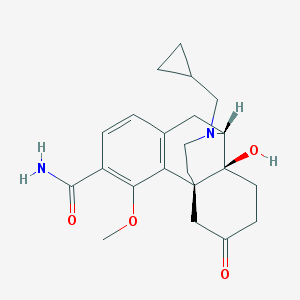 CHEMBL195285 CHEMBL195285 | C22H28N2O4 | 384.476 | 5 / 2 | 0.9 | Yes |
| 6062 | 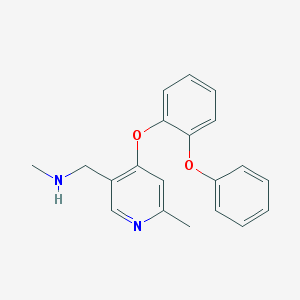 CHEMBL273208 CHEMBL273208 | C20H20N2O2 | 320.392 | 4 / 1 | 3.7 | Yes |
| 6174 |  CHEMBL3262366 CHEMBL3262366 | C34H43NO4 | 529.721 | 5 / 2 | 5.8 | No |
| 6181 | 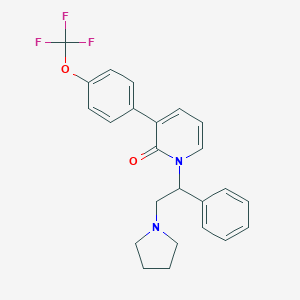 CHEMBL61426 CHEMBL61426 | C24H23F3N2O2 | 428.455 | 6 / 0 | 5.3 | No |
| 6402 | 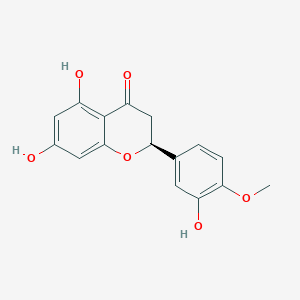 hesperetin hesperetin | C16H14O6 | 302.282 | 6 / 3 | 2.4 | Yes |
| 441936 | 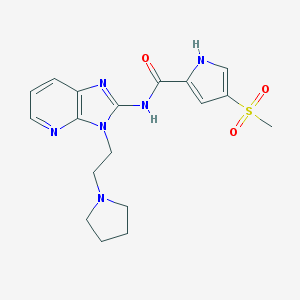 CHEMBL3394008 CHEMBL3394008 | C18H22N6O3S | 402.473 | 6 / 2 | 0.7 | Yes |
| 6665 |  CHEMBL265195 CHEMBL265195 | C28H29NO10 | 539.537 | 10 / 0 | 4.0 | No |
| 6786 |  spiropiperidine analogue, 11 spiropiperidine analogue, 11 | C28H31N3O | 425.576 | 3 / 0 | 5.4 | No |
| 463682 |  CHEMBL3597076 CHEMBL3597076 | C83H134N20O26 | 1828.1 | 29 / 25 | -6.6 | No |
| 6813 |  CHEMBL41098 CHEMBL41098 | C38H42N4O8S2 | 746.894 | 12 / 4 | 2.1 | No |
| 6817 |  KUC104543N KUC104543N | C26H30N6O2 | 458.566 | 5 / 1 | 2.7 | Yes |
| 521646 | 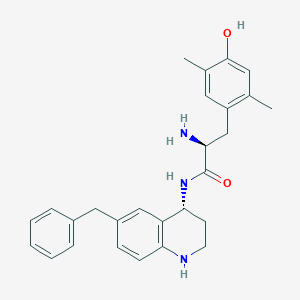 CHEMBL3770677 CHEMBL3770677 | C27H31N3O2 | 429.564 | 4 / 4 | 4.6 | Yes |
| 6920 | 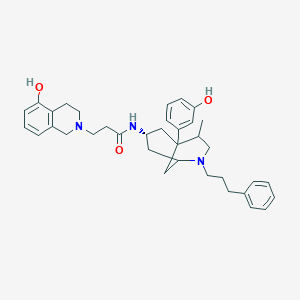 CHEMBL382597 CHEMBL382597 | C36H45N3O3 | 567.774 | 5 / 3 | 6.0 | No |
| 6936 | 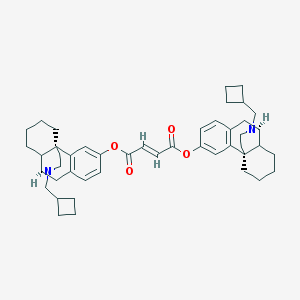 MCL-145 MCL-145 | C46H58N2O4 | 702.98 | 6 / 0 | 10.6 | No |
| 6937 | 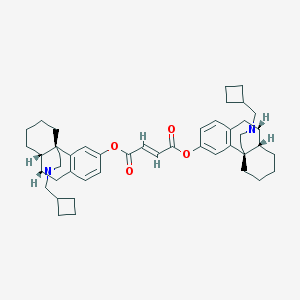 CHEMBL146756 CHEMBL146756 | C46H58N2O4 | 702.98 | 6 / 0 | 10.6 | No |
| 463705 | 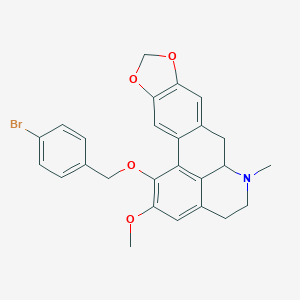 CHEMBL3633757 CHEMBL3633757 | C26H24BrNO4 | 494.385 | 5 / 0 | 5.4 | No |
| 6992 | 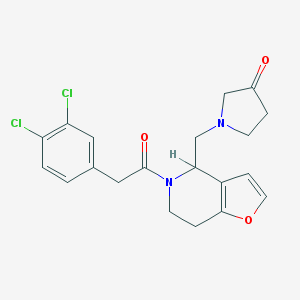 CHEMBL70226 CHEMBL70226 | C20H20Cl2N2O3 | 407.291 | 4 / 0 | 2.8 | Yes |
| 7008 | 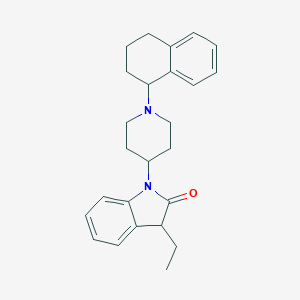 CHEMBL3262546 CHEMBL3262546 | C25H30N2O | 374.528 | 2 / 0 | 4.8 | Yes |
| 7011 | 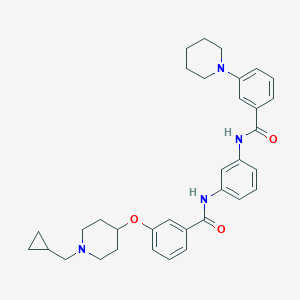 CHEMBL2322511 CHEMBL2322511 | C34H40N4O3 | 552.719 | 5 / 2 | 5.9 | No |
| 557482 | 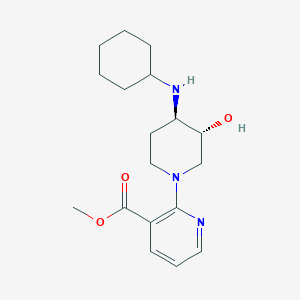 SCHEMBL16944666 SCHEMBL16944666 | C18H27N3O3 | 333.432 | 6 / 2 | 2.1 | Yes |
| 463757 |  CHEMBL3183897 CHEMBL3183897 | C18H27N3O3 | 333.432 | 6 / 2 | 2.1 | Yes |
| 521655 | 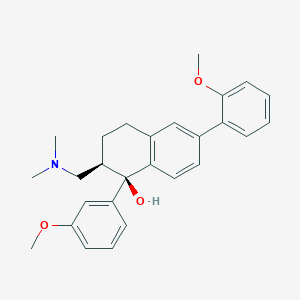 CHEMBL3808879 CHEMBL3808879 | C27H31NO3 | 417.549 | 4 / 1 | 4.8 | Yes |
| 557486 |  SCHEMBL15486274 SCHEMBL15486274 | C32H47NO7 | 557.728 | 8 / 0 | 3.4 | No |
| 547971 | 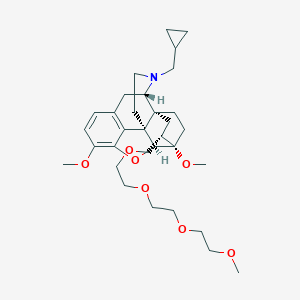 CHEMBL3959095 CHEMBL3959095 | C32H47NO7 | 557.728 | 8 / 0 | 3.4 | No |
| 7109 | 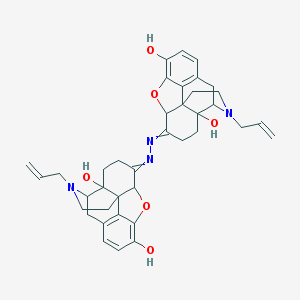 NSC612113 NSC612113 | C38H42N4O6 | 650.776 | 10 / 4 | 4.0 | No |
| 7112 | 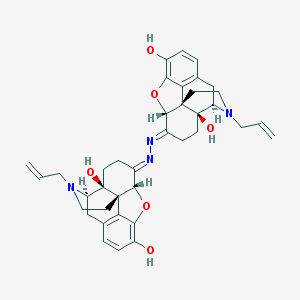 Naloxonazine Naloxonazine | C38H42N4O6 | 650.776 | 10 / 4 | 4.0 | No |
| 7132 | 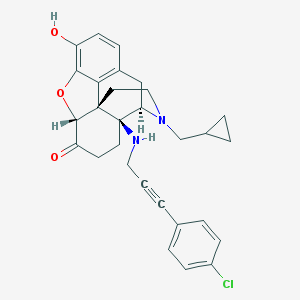 CHEMBL574175 CHEMBL574175 | C29H29ClN2O3 | 489.012 | 5 / 2 | 4.7 | Yes |
| 557489 | 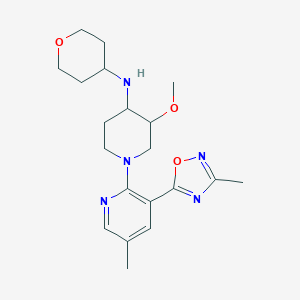 SCHEMBL16944858 SCHEMBL16944858 | C20H29N5O3 | 387.484 | 8 / 1 | 2.0 | Yes |
| 557496 | 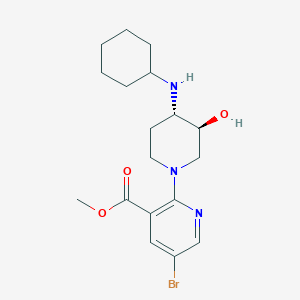 SCHEMBL16945084 SCHEMBL16945084 | C18H26BrN3O3 | 412.328 | 6 / 2 | 2.8 | Yes |
| 7330 | 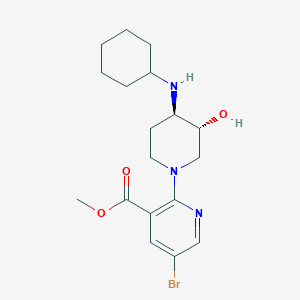 CHEMBL3264446 CHEMBL3264446 | C18H26BrN3O3 | 412.328 | 6 / 2 | 2.8 | Yes |
| 463785 |  MLS004801944 MLS004801944 | C18H26BrN3O3 | 412.328 | 6 / 2 | 2.8 | Yes |
| 7486 | 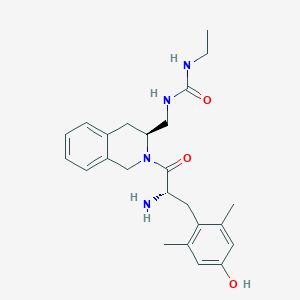 CHEMBL309783 CHEMBL309783 | C24H32N4O3 | 424.545 | 4 / 4 | 2.3 | Yes |
| 536163 | 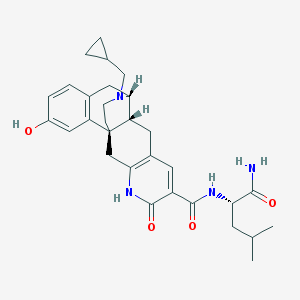 CHEMBL3956104 CHEMBL3956104 | C30H38N4O4 | 518.658 | 5 / 4 | 3.4 | No |
| 7580 | 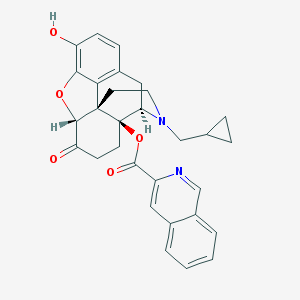 CHEMBL516085 CHEMBL516085 | C30H28N2O5 | 496.563 | 7 / 1 | 4.7 | Yes |
| 7627 |  CHEMBL413258 CHEMBL413258 | C31H31N3O6 | 541.604 | 7 / 1 | 4.0 | No |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218