| # |
Accession |
Prediction with UniProt 2018_02 template database |
Matcha,b 2018_02 |
Fmax MF/BP Match 2018_02 |
Prediction with UniProt 2018_11 template database |
Matcha,b 2018_11 |
Fmax MF/BP Match 2018_11 |
neXtProt annotation |
Explanation of change of our predictionc |
| 1 |
O75363-1 |
(for CC: neuron part)
 |
|
NA,0.40 |
neural cell part, such as myelin
 |
* |
NA/0.80 |
Required for myelination. |
Earlier prediction used information in UniProt_11, which included experimental annotations of target proteins |
| 2 |
O75677-1 |
ubiquitin-protein transferase activity
 |
* |
NA,0.27 |
ubiquitin-protein transferase activity
 |
* |
NA/0.22 |
Negatively regulates the G2-M phase transition, possibly by promoting cyclin B1/CCNB1 and CDK1 proteasomal degradation and thereby preventing their accumulation during interphase. |
|
| 3 |
P0C870-1 |
histone demethylation
 |
* |
0.55,0.90 |
oxidoreductase for oxidative demethylation
 |
* |
0.71/NA |
Bifunctional enzyme that acts both as an endopeptidase and 2-oxoglutarate-dependent monoxygenase. Endopeptidase that cleaves histones N-terminal tails at the carboxyl side of methylated arginine or lysine residues, to generate 'tailless nucleosomes', which may trigger transcription elongation. Preferentially recognizes and cleaves monomethylated and dimethylated arginine residues of histones H2, H3 and H4. After initial cleavage, continues to digest histones tails via its aminopeptidase activity. Additionally, may play a role in protein biosynthesis by modifying the translation machinery. Acts as Fe2+ and 2-oxoglutarate-dependent monoxygenase, catalyzing (S)-stereospecific hydroxylation at C-3 of 'Lys-22' of DRG1 and 'Lys-21' of DRG2 translation factors (TRAFAC), promoting their interaction with ribonucleic acids (RNA). |
|
| 4 |
P60827-1 |
signaling receptor binding
 |
* |
NA,0.40 |
extracellular matrix structure
 |
|
NA/0.36 |
May play a role as ligand of relaxin receptor RXFP1. |
While the corrected prediction is not exactly correct (as the earlier prediction), it suggests signaling receptor binding. Since the protein is binds to signal receptor RXFP1, the corrected prediction is roughly consistent with the earlier prediction. |
| 5 |
Q494U1-1 |
transmembrane transport of small molecules, such as nucleotide
 |
|
0.05,0.29 |
transmembrane transport of small molecules, such as nucleotide
 |
+ |
0.58/0.78 |
Controls the stability of the leptin mRNA harboring an AU-rich element (ARE) in its 3' UTR, in cooperation with the RNA stabilizer ELAVL1 |
|
| 6 |
Q5T0D9-1 |
phosphatidylinositol-4-phosphate phosphatase
 |
+ |
NA,0.55 |
phosphatidylinositol-4-phosphate phosphatase
 |
|
NA/0.21 |
Presynaptic protein involved in the synaptic transmission tuning. Regulates synaptic release probability by decreasing the calcium sensitivity of release |
|
| 7 |
Q5VTQ0-1 |
protein ubiquitination regulation
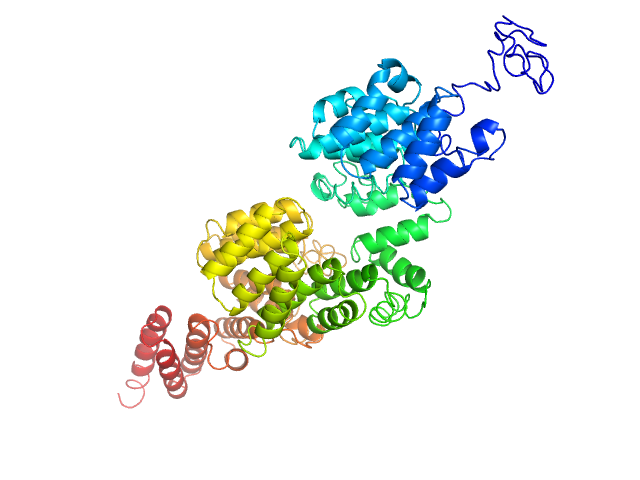 |
* |
NA,0.26 |
cholesterol metabolism
 |
* |
NA/0.94 |
Regulates high density lipoprotein (HDL) cholesterol metabolism by promoting the ubiquitination and degradation of the oxysterols receptors LXR (NR1H2 and NR1H3). |
The corrected prediction can predict ubiquitination but not the cholesterol metabolism. Since only part of the functions are correctly predicted, the Fmax is low. |
| 8 |
Q6AI39-1 |
sodium:potassium ion transporter
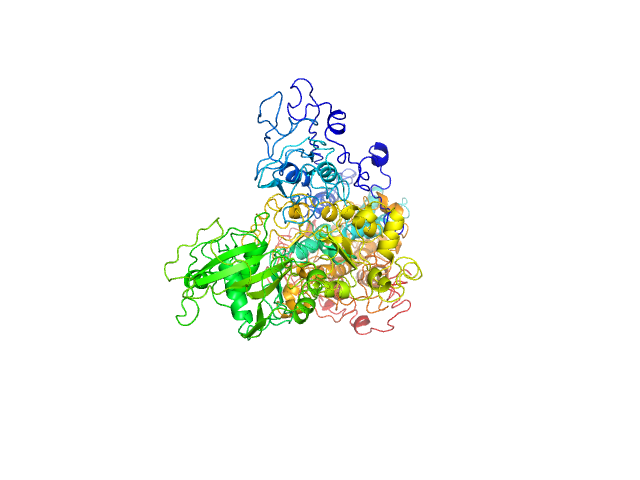 |
|
NA,NA |
regulation of expression at chromosome level
 |
* |
NA/NA |
Component of SWI/SNF chromatin remodeling subcomplex GBAF that carries out key enzymatic activities, changing chromatin structure by altering DNA-histone contacts within a nucleosome in an ATP-dependent manner. |
After removing the bias in the earlier prediction, the corrected prediction can not predict the specific function due to lack of template information. |
| 9 |
Q6ZNE9-2 |
regulation of protein folding
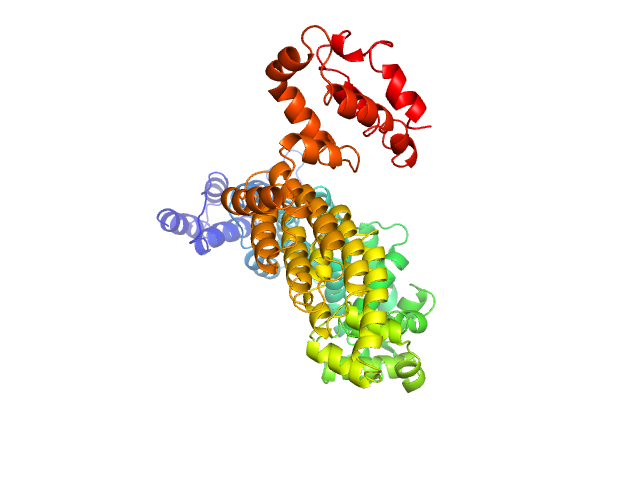 |
|
0.26,0.32 |
nucleobase-containing compound biosynthetic process
 |
|
0.35/0.41 |
Positively regulates macroautophagy in primary dendritic cells. Increases autophagic flux, probably by stimulating both autophagosome formation and facilitating tethering with lysosomes. Binds to phosphatidylinositol 3-phosphate (PtdIns3P) through its FYVE-type zinc finger. |
|
| 10 |
Q7Z5A7-1 |
regulation of microglial cell activation
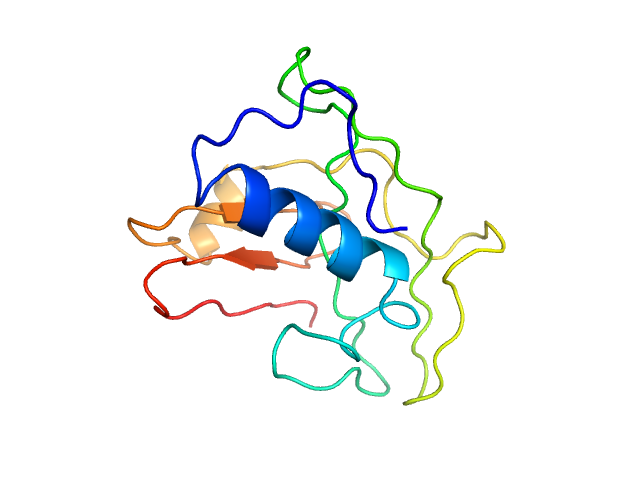 |
* |
0.67,0.50 |
oxidoreductase
 |
+ |
0.62/0.46 |
Acts as a chemokine-like protein by regulating cell proliferation and migration through activation of G protein-coupled receptors (GPCRs), such as S1PR2 and FPR2. Stimulates chemotactic migration of macrophages mediated by the MAPK3/ERK1 and AKT1 pathway. Blocks TNFSF11/RANKL-induced osteoclast formation from macrophages by inhibiting up-regulation of osteoclast fusogenic and differentiation genes. Stimulation of macrophage migration and inhibition of osteoclast formation is mediated via GPCR FPR2. Acts as an adipokine by negatively regulating vascular smooth muscle cell (VSMC) proliferation and migration in response to platelet-derived growth factor stimulation via GPCR S1PR2 and G protein GNA12/GNA13-transmitted RHOA signaling. Inhibits injury-induced cell proliferation and neointima formation in the femoral arteries |
The corrected prediction includes more specific function for activation of microglial cells (macrophages in central nerve system). |
| 11 |
Q8IUW5-1 |
regulation of apoptosis through TNF
 |
* |
NA,0.29 |
p38 MAPK cascade regulation
 |
* |
NA/1.00 |
Induces activation of MAPK14/p38 cascade, when overexpressed |
While the corrected prediction cannot specifically predict p38/MAPK cascade regulation, the corrected prediction suggest that this protein regulate apoptosis through TNF. Since MAPK is known to regulate TNF-mediated apoptosis, the prediction is roughly consistent with neXtProt annotation. |
| 12 |
Q8IUY3-1 |
binding of GTPase from Ras superfamily
 |
|
0.11,0.38 |
Binding of GTPase from Ras superfamily
 |
|
0.36/0.41 |
Participates in the organization of endoplasmic reticulum-plasma membrane contact sites (EPCS) with pleiotropic functions including STIM1 recruitment and calcium homeostasis. Constitutive tether that co-localize with ESYT2/3 tethers at endoplasmic reticulum-plasma membrane contact sites in a phosphatidylinositol lipid-dependent manner. Pre-marks the subset of phosphtidylinositol 4,5-biphosphate (PI(4,5)P2)-enriched EPCS destined for the store operated calcium entry pathway (SOCE). |
|
| 13 |
Q8NDM7-1 |
 |
|
NA,0.29 |
ciliary structure and movement
 |
* |
NA/0.45 |
Flagellar protein involved in sperm flagellum axoneme organization and function. |
After removing the bias in the earlier prediction, the corrected prediction can not predict the specific function due to lack of template information. |
| 14 |
Q8TDG2-1 |
regulation of chromosome organization either though histone acetylation or binding of cytoskelton used in chromsome segregation
 |
|
0.03,0.29 |
structure component of cytoskeleton
 |
|
0.04/0.40 |
Negatively regulates the Hedgehog (SHH) signaling. Binds to the promoter of the SHH signaling mediator, GLI1, and inhibits its expression. |
|
| 15 |
Q8WTR8-1 |
anatomical structure morphogenesis
 |
* |
NA,0.44 |
anatomical structure morphogenesis
 |
* |
NA/0.40 |
Plays a role in neurogenesis. Prevents motor neuron cell body migration out of the neural tube. |
|
| 16 |
Q96D15-1 |
catalytic activity, acting on a protein
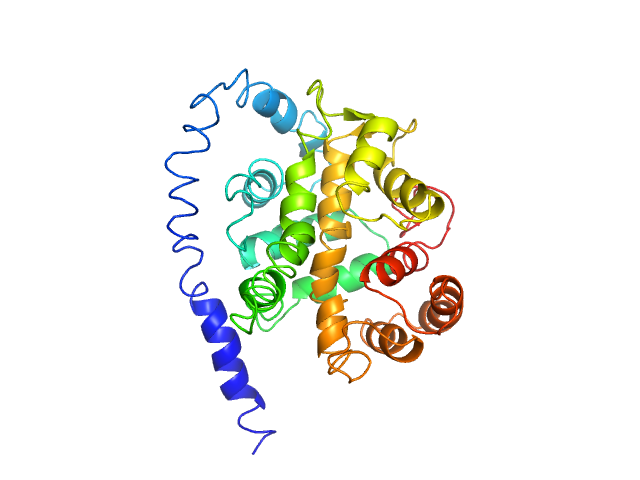 |
|
NA,0.47 |
 |
|
NA/0.48 |
Probable molecular chaperone assisting protein biosynthesis and transport in the endoplasmic reticulum. Required for the proper biosynthesis and transport of pulmonary surfactant-associated protein A/SP-A, pulmonary surfactant-associated protein D/SP-D and the lipid transporter ABCA3. By regulating both the proper expression and the degradation through the endoplasmic reticulum-associated protein degradation pathway of these proteins plays a crucial role in pulmonary surfactant homeostasis. Has an anti-fibrotic activity by negatively regulating the secretion of type I and type III collagens. This calcium-binding protein also transiently associates with immature PCSK6 and regulates its secretion. |
|
| 17 |
Q96J88-1 |
cytoskeleton binding
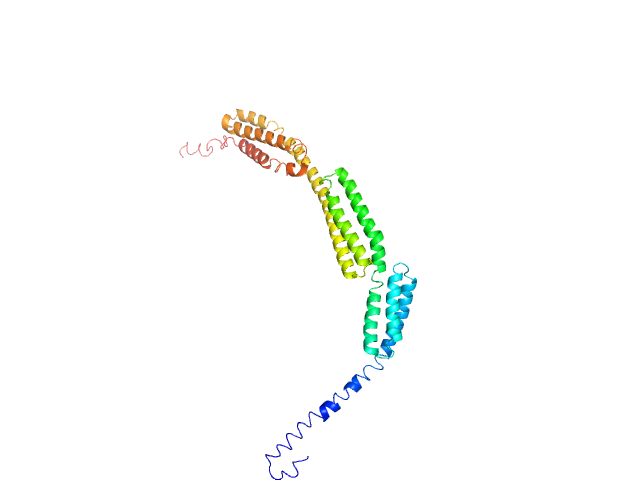 |
|
NA,NA |
cytoskelton binding
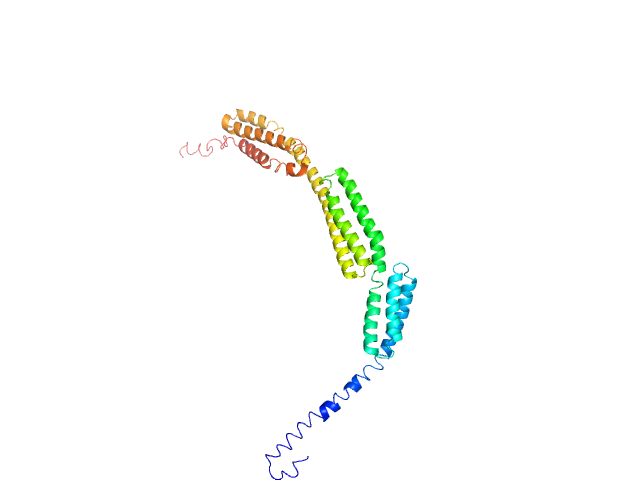 |
|
NA/NA |
Plays a role in M1 macrophage polarization and is required for the proper regulation of gene expression during M1 versus M2 macrophage differentiation. Might play a role in RELA/p65 and STAT1 phosphorylation and nuclear localization upon activation of macrophages. |
|
| 18 |
Q96KV7-1 |
regulation of transcription by nucleic acid binding
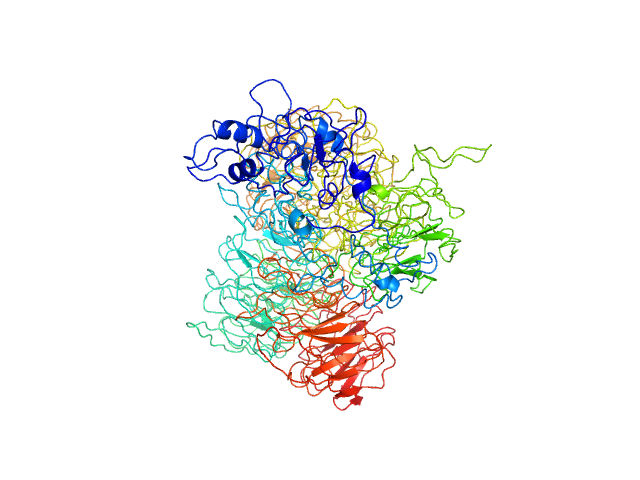 |
|
NA,0.17 |
regulation of transcription by nucleic acid binding
 |
|
NA/0.18 |
Required for efficient primary cilium formation. |
|
| 19 |
Q96M27-1 |
protein kinase A regulation
 |
* |
1.00,0.88 |
protein kinase A regulation
 |
* |
1.00/0.92 |
Activation of protein kinase A activity. Protein binding. Protein kinase A regulatory subunit binding. |
No change. In both versions of our prediction, we only used the zebrafish ortholog annotation, which is in the UniProt database since 2011. Since we did not use function annotation of the mammalian ortholog as template during prediction, this is not a bias of our approach. |
| 20 |
Q96S16-1 |
histone demethylation
 |
|
NA,0.21 |
oxidoreductase for oxidative demethylation
 |
+ |
NA/0.57 |
Functions as a positive regulator of TNF-induced NF-kappa-B signaling. Regulates angiogenesis and cellular metabolism through interaction with PKM. |
|
| 21 |
Q9BZD6-1 |
serine-type endopeptidase
 |
|
NA,NA |
serine-type endopeptidase
 |
|
NA/NA |
May control axon guidance across the CNS. Prevents the delivery of ROBO1 at the cell surface and downregulates its expression. |
|
| 22 |
Q9BZH6-1 |
 |
|
NA,0.35 |
 |
+ |
NA/0.80 |
Involved in the Hedgehog (Hh) signaling pathway, is essential for normal ciliogenesis. Regulates the proteolytic processing of GLI3 and cooperates with the transcription factor EMX1 in the induction of downstream Hh pathway gene expression and gonadotropin-releasing hormone production. WDR11 complex facilitates the tethering of Adaptor protein-1 complex (AP-1)-derived vesicles. WDR11 complex acts together with TBC1D23 to facilitate the golgin-mediated capture of vesicles generated using AP-1. |
|
| 23 |
Q9C0D6-1 |
binding of cytoskeleton
 |
* |
0.44,0.33 |
binding of cytoskeleton, such as actin
 |
* |
0.67/0.43 |
Microtubule-associated formin which regulates both actin and microtubule dynamics. Induces microtubule acetylation and stabilization and actin stress fiber formation. Regulates Golgi ribbon formation. Required for normal cilia assembly. |
|
| 24 |
Q9GZU8-1 |
hydrolase, probably hydrolase of protein
 |
|
NA,0.32 |
hydrolase, probably hydrolase of proteins
 |
|
NA/0.33 |
Promotes the association of the proteasome activator complex subunit PSME3 with the 20S proteasome and regulates its activity. Inhibits PSME3-mediated degradation of some proteasome substrates, probably by affecting their diffusion rate into the catalytic chamber of the proteasome |
While this protein is related to protein hydrolysis, it is not a hydrolase itself. |
| 25 |
Q9H9L7-1 |
by binding to RNA polymerase, regulate expression of genes such as cytokines
 |
|
NA,0.18 |
repress transcription by binding to RNA polymerase
 |
|
NA/0.18 |
Functions as signal transducer for MSTN during skeletal muscle regeneration and myogenesis. May regulates chemotaxis of both macrophages and myoblasts by reorganising actin cytoskeleton, leading to more efficient lamellipodia formation via a PI3 kinase dependent pathway. |
|