

You can:
| Name | Extracellular calcium-sensing receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | CASR |
| Synonym | hCasR {ECO:0000303|PubMed:27386547} GPRC2A extracellular calcium-sensing receptor Parathyroid cell calcium-sensing receptor 1 {ECO:0000303|PubMed:8698326} PCaR1 {ECO:0000303|PubMed:8698326} [ Show all ] |
| Disease | Osteoporosis Cerebrovascular ischaemia Cardiovascular disease; Kidney disease Secondary hyperparathyroidism Myelodysplastic syndrome [ Show all ] |
| Length | 1078 |
| Amino acid sequence | MAFYSCCWVLLALTWHTSAYGPDQRAQKKGDIILGGLFPIHFGVAAKDQDLKSRPESVECIRYNFRGFRWLQAMIFAIEEINSSPALLPNLTLGYRIFDTCNTVSKALEATLSFVAQNKIDSLNLDEFCNCSEHIPSTIAVVGATGSGVSTAVANLLGLFYIPQVSYASSSRLLSNKNQFKSFLRTIPNDEHQATAMADIIEYFRWNWVGTIAADDDYGRPGIEKFREEAEERDICIDFSELISQYSDEEEIQHVVEVIQNSTAKVIVVFSSGPDLEPLIKEIVRRNITGKIWLASEAWASSSLIAMPQYFHVVGGTIGFALKAGQIPGFREFLKKVHPRKSVHNGFAKEFWEETFNCHLQEGAKGPLPVDTFLRGHEESGDRFSNSSTAFRPLCTGDENISSVETPYIDYTHLRISYNVYLAVYSIAHALQDIYTCLPGRGLFTNGSCADIKKVEAWQVLKHLRHLNFTNNMGEQVTFDECGDLVGNYSIINWHLSPEDGSIVFKEVGYYNVYAKKGERLFINEEKILWSGFSREVPFSNCSRDCLAGTRKGIIEGEPTCCFECVECPDGEYSDETDASACNKCPDDFWSNENHTSCIAKEIEFLSWTEPFGIALTLFAVLGIFLTAFVLGVFIKFRNTPIVKATNRELSYLLLFSLLCCFSSSLFFIGEPQDWTCRLRQPAFGISFVLCISCILVKTNRVLLVFEAKIPTSFHRKWWGLNLQFLLVFLCTFMQIVICVIWLYTAPPSSYRNQELEDEIIFITCHEGSLMALGFLIGYTCLLAAICFFFAFKSRKLPENFNEAKFITFSMLIFFIVWISFIPAYASTYGKFVSAVEVIAILAASFGLLACIFFNKIYIILFKPSRNTIEEVRCSTAAHAFKVAARATLRRSNVSRKRSSSLGGSTGSTPSSSISSKSNSEDPFPQPERQKQQQPLALTQQEQQQQPLTLPQQQRSQQQPRCKQKVIFGSGTVTFSLSFDEPQKNAMAHRNSTHQNSLEAQKSSDTLTRHEPLLPLQCGETDLDLTVQETGLQGPVGGDQRPEVEDPEELSPALVVSSSQSFVISGGGSTVTENVVNS |
| UniProt | P41180 |
| Protein Data Bank | 5k5t, 5k5s, 5fbk, 5fbh |
| GPCR-HGmod model | N/A |
| 3D structure model | This structure is from PDB ID 5k5t. |
| BioLiP | BL0353424,BL0353430, BL0353434, BL0353425,BL0353426,BL0353427,, BL0349612,BL0349615, BL0349611,BL0349614, BL0349610,BL0349613, BL0349606,BL0349609, BL0349605,BL0349608, BL0349604,BL0349607 |
| Therapeutic Target Database | T92076 |
| ChEMBL | CHEMBL1878 |
| IUPHAR | 54 |
| DrugBank | BE0000509 |
| Name | CHEMBL200312 |
|---|---|
| Molecular formula | C26H31N3OS |
| IUPAC name | 3-[[(2R)-2-[(1R)-2-[[1-(1-benzothiophen-2-yl)-2-methylpropan-2-yl]amino]-1-hydroxyethyl]pyrrolidin-1-yl]methyl]benzonitrile |
| Molecular weight | 433.614 |
| Hydrogen bond acceptor | 5 |
| Hydrogen bond donor | 2 |
| XlogP | 4.6 |
| Synonyms | GTPL711 3-{[(2R)-2-[(1R)-2-{[1-(1-benzothiophen-2-yl)-2-methylpropan-2-yl]amino}-1-hydroxyethyl]pyrrolidin-1-yl]methyl}benzonitrile BDBM50175363 compound 26a [PMID: 16216508] 1-arylmethylpyrrolidin-2-yl ethanol amine [ Show all ] |
| Inchi Key | HCLQARMRCPEALF-DNQXCXABSA-N |
| Inchi ID | InChI=1S/C26H31N3OS/c1-26(2,15-22-14-21-9-3-4-11-25(21)31-22)28-17-24(30)23-10-6-12-29(23)18-20-8-5-7-19(13-20)16-27/h3-5,7-9,11,13-14,23-24,28,30H,6,10,12,15,17-18H2,1-2H3/t23-,24-/m1/s1 |
| PubChem CID | 44405854 |
| ChEMBL | CHEMBL200312 |
| IUPHAR | 711 |
| BindingDB | 50175363 |
| DrugBank | N/A |
Structure | 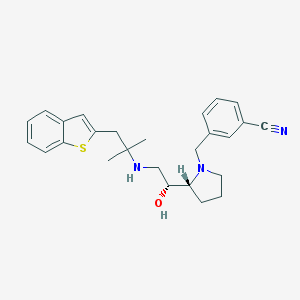 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| IC50 | 270.0 nM | PMID16216508 | BindingDB,ChEMBL |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218