

You can:
| Name | Adenosine receptor A1 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | ADORA1 |
| Synonym | RDC7 A1 receptor A1-AR A1R adenosine receptor A1 |
| Disease | Cardiac arrhythmias Hypertension Cardiac disease Cognitive disorders Diabetes [ Show all ] |
| Length | 326 |
| Amino acid sequence | MPPSISAFQAAYIGIEVLIALVSVPGNVLVIWAVKVNQALRDATFCFIVSLAVADVAVGALVIPLAILINIGPQTYFHTCLMVACPVLILTQSSILALLAIAVDRYLRVKIPLRYKMVVTPRRAAVAIAGCWILSFVVGLTPMFGWNNLSAVERAWAANGSMGEPVIKCEFEKVISMEYMVYFNFFVWVLPPLLLMVLIYLEVFYLIRKQLNKKVSASSGDPQKYYGKELKIAKSLALILFLFALSWLPLHILNCITLFCPSCHKPSILTYIAIFLTHGNSAMNPIVYAFRIQKFRVTFLKIWNDHFRCQPAPPIDEDLPEERPDD |
| UniProt | P30542 |
| Protein Data Bank | 6d9h, 5n2s |
| GPCR-HGmod model | P30542 |
| 3D structure model | This structure is from PDB ID 6d9h. |
| BioLiP | BL0385576, BL0417675 |
| Therapeutic Target Database | T88714, T92072 |
| ChEMBL | CHEMBL226 |
| IUPHAR | 18 |
| DrugBank | BE0000013 |
| Name | NECA |
|---|---|
| Molecular formula | C12H16N6O4 |
| IUPAC name | (2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-N-ethyl-3,4-dihydroxyoxolane-2-carboxamide |
| Molecular weight | 308.298 |
| Hydrogen bond acceptor | 8 |
| Hydrogen bond donor | 4 |
| XlogP | -0.7 |
| Synonyms | (2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-N-ethyl-3,4-dihydroxyoxolane-2-carboxamide 5''-N-Ethylcarboxamidoadenosine; D-NECA; NECAMolecular Formula: C12H16N6O4 74992-42-0 Adenosine-5-(N-ethylcarboxamide) CS-8131 [ Show all ] |
| Inchi Key | JADDQZYHOWSFJD-FLNNQWSLSA-N |
| Inchi ID | InChI=1S/C12H16N6O4/c1-2-14-11(21)8-6(19)7(20)12(22-8)18-4-17-5-9(13)15-3-16-10(5)18/h3-4,6-8,12,19-20H,2H2,1H3,(H,14,21)(H2,13,15,16)/t6-,7+,8-,12+/m0/s1 |
| PubChem CID | 448222 |
| ChEMBL | CHEMBL464859 |
| IUPHAR | 377, 425 |
| BindingDB | 21220 |
| DrugBank | N/A |
Structure | 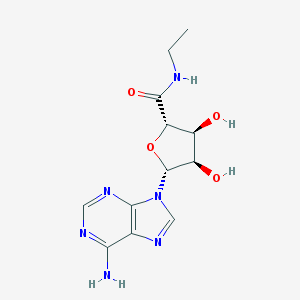 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| Activity | -73.4 % | PMID26756468 | ChEMBL |
| Activity | 73.0 % | MedChemComm, (2012) 3:3:333 | ChEMBL |
| Activity | 177.0 % | MedChemComm, (2012) 3:3:333 | ChEMBL |
| Affinity constant | 0.1 uM | PMID6279840 | ChEMBL |
| EC50 | 0.3631 nM | PMID17249651 | ChEMBL |
| EC50 | 1.259 nM | MedChemComm, (2014) 5:2:192 | ChEMBL |
| EC50 | 10.2 nM | PMID21858244 | BindingDB,ChEMBL |
| EC50 | 30.0 nM | PMID20031406 | BindingDB,ChEMBL |
| EC50 | 31.62 nM | Med Chem Res, (2004) 13:1:88 | ChEMBL |
| EC50 | 59.0 nM | PMID9871584 | BindingDB,ChEMBL |
| EC50 | 1000.0 nM | PMID26756468 | BindingDB,ChEMBL |
| EC50 | 1479.11 nM | MedChemComm, (2012) 3:3:333 | ChEMBL |
| Efficacy | 100.0 % | PMID22486652 | ChEMBL |
| IC50 | 0.2089 nM | PMID26756468 | ChEMBL |
| IC50 | 0.20893 nM | PMID26756468 | BindingDB |
| IC50 | 0.7943 nM | MedChemComm, (2012) 3:3:333 | ChEMBL |
| Inhibition | 83.3 % | PMID17249651 | ChEMBL |
| Inhibition | 98.0 % | PMID17249651 | ChEMBL |
| Inhibition | 100.0 % | PMID21858244, PMID22921089 | ChEMBL |
| Inhibition | 105.0 % | MedChemComm, (2012) 3:3:333 | ChEMBL |
| Kd | 8.6 nM | PMID15771445 | BindingDB,ChEMBL |
| Kd | 575.0 nM | PMID17249651 | BindingDB |
| Kd | 575.44 nM | PMID17249651 | ChEMBL |
| KD hydro | 0.25 mM | PMID15771445 | ChEMBL |
| Ki | <200.0 nM | PMID11714602 | BindingDB,ChEMBL |
| Ki | 1.04 nM | PMID16487705 | BindingDB,ChEMBL |
| Ki | 3.0 nM | PMID22486652 | BindingDB,ChEMBL |
| Ki | 5.0 nM | PMID15267242 | BindingDB,ChEMBL |
| Ki | 6.30957 - 5011.87 nM | PMID7798201, PMID9920910, PMID8300561, PMID15476669, PMID14662005 | IUPHAR |
| Ki | 6.8 nM | PMID27933810, PMID15734651, PMID22921089, PMID17378544 | BindingDB,ChEMBL |
| Ki | 7.8 nM | PMID18637670 | BindingDB,ChEMBL |
| Ki | 8.6 nM | PMID25780876 | BindingDB |
| Ki | 8.62 nM | PMID25780876 | ChEMBL |
| Ki | 12.0 nM | PMID12672250, PMID15239649 | BindingDB,ChEMBL |
| Ki | 13.6 nM | PMID24900277, PMID23245803, PMID10212124 | BindingDB,ChEMBL |
| Ki | 14.0 nM | PMID17927167, PMID20408530, PMID23245803, PMID10212124, PMID10494877, PMID24077183, PMID24164628, PMID22257095, PMID11459663, PMID16366607, PMID18269230 | BindingDB,ChEMBL |
| Ki | 18.2 nM | PMID17306548, PMID15481989 | BindingDB,ChEMBL |
| Ki | 18.3 nM | PMID17228880 | BindingDB,ChEMBL |
| Ki | 1122.02 nM | MedChemComm, (2012) 3:3:333 | ChEMBL |
| pKA | 4.4 - | PMID26756468 | ChEMBL |
| Ratio | 3.2 - | PMID17249651 | ChEMBL |
| Relative potency | 0.2 - | PMID12672250 | ChEMBL |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218