

You can:
| Name | Cannabinoid receptor 2 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | CNR2 |
| Synonym | Peripheral cannabinoid receptor rCB2 hCB2 cannabinoid receptor 2 (macrophage) cannabinoid receptor 2 (spleen) [ Show all ] |
| Disease | Immune disorder Inflammatory bowel disease Inflammatory disease Neuropathic pain Osteoporosis [ Show all ] |
| Length | 360 |
| Amino acid sequence | MEECWVTEIANGSKDGLDSNPMKDYMILSGPQKTAVAVLCTLLGLLSALENVAVLYLILSSHQLRRKPSYLFIGSLAGADFLASVVFACSFVNFHVFHGVDSKAVFLLKIGSVTMTFTASVGSLLLTAIDRYLCLRYPPSYKALLTRGRALVTLGIMWVLSALVSYLPLMGWTCCPRPCSELFPLIPNDYLLSWLLFIAFLFSGIIYTYGHVLWKAHQHVASLSGHQDRQVPGMARMRLDVRLAKTLGLVLAVLLICWFPVLALMAHSLATTLSDQVKKAFAFCSMLCLINSMVNPVIYALRSGEIRSSAHHCLAHWKKCVRGLGSEAKEEAPRSSVTETEADGKITPWPDSRDLDLSDC |
| UniProt | P34972 |
| Protein Data Bank | 5zty |
| GPCR-HGmod model | P34972 |
| 3D structure model | This structure is from PDB ID 5zty. |
| BioLiP | BL0438927 |
| Therapeutic Target Database | T37693 |
| ChEMBL | CHEMBL253 |
| IUPHAR | 57 |
| DrugBank | BE0000095 |
| Name | Rimonabant |
|---|---|
| Molecular formula | C22H21Cl3N4O |
| IUPAC name | 5-(4-chlorophenyl)-1-(2,4-dichlorophenyl)-4-methyl-N-piperidin-1-ylpyrazole-3-carboxamide |
| Molecular weight | 463.787 |
| Hydrogen bond acceptor | 3 |
| Hydrogen bond donor | 1 |
| XlogP | 6.5 |
| Synonyms | Zimulti D0E0ZR EX-A688 J-010440 143881-EP2292620A2 [ Show all ] |
| Inchi Key | JZCPYUJPEARBJL-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C22H21Cl3N4O/c1-14-20(22(30)27-28-11-3-2-4-12-28)26-29(19-10-9-17(24)13-18(19)25)21(14)15-5-7-16(23)8-6-15/h5-10,13H,2-4,11-12H2,1H3,(H,27,30) |
| PubChem CID | 104850 |
| ChEMBL | CHEMBL111 |
| IUPHAR | 743 |
| BindingDB | 21278 |
| DrugBank | DB06155 |
Structure | 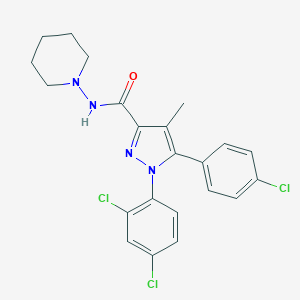 |
| Lipinski's druglikeness | This ligand has a partition coefficient log P greater than 5. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| EC50 | 31210.0 nM | PMID19328683 | BindingDB,ChEMBL |
| Efficacy | 0.0 % | PMID24900561 | ChEMBL |
| Emax | 100.0 % | PMID26151231 | ChEMBL |
| IC50 | <1000.0 nM | PMID19128970 | BindingDB,ChEMBL |
| IC50 | 1.9 nM | PMID19095444 | BindingDB,ChEMBL |
| IC50 | 92.5 nM | PMID19850473 | BindingDB,ChEMBL |
| IC50 | 600.0 nM | PMID17181138 | BindingDB,ChEMBL |
| IC50 | 603.3 nM | PMID17293109 | BindingDB,ChEMBL |
| IC50 | 1631.1 nM | PMID19530697 | BindingDB,ChEMBL |
| IC50 | 1760.0 nM | PMID19022666, PMID18954042, PMID20045337, PMID19850473, PMID18337096, PMID20673729, PMID19269817, PMID18243711 | BindingDB,ChEMBL |
| IC50 | 1939.8 nM | PMID18712856 | BindingDB,ChEMBL |
| IC50 | 1980.0 nM | PMID21741835 | BindingDB,ChEMBL |
| IC50 | 4570.0 nM | PMID18754613 | BindingDB,ChEMBL |
| IC50 | 9100.0 nM | PMID26151231 | BindingDB,ChEMBL |
| IC50 | 9800.0 nM | PMID25644673, PMID24445310 | BindingDB,ChEMBL |
| Inhibition | 0.0 % | PMID22916707 | ChEMBL |
| Inhibition | 9.0 % | PMID25096297 | ChEMBL |
| Inhibition | 34.0 % | PMID21741835 | ChEMBL |
| Inhibition | 38.6 % | PMID24445310 | ChEMBL |
| Inhibition | 100.0 % | PMID24445310 | ChEMBL |
| Kb | <1977.0 nM | PMID18800770 | ChEMBL |
| Ki | <1000.0 nM | PMID18363352, PMID19520572 | BindingDB,ChEMBL |
| Ki | 1.1 nM | PMID24856183 | BindingDB,ChEMBL |
| Ki | 126.0 nM | PMID20047779 | PDSP,BindingDB,ChEMBL |
| Ki | 313.0 nM | PMID22372835, PMID26827137, PMID12061874, PMID20845959, PMID18512901 | PDSP,BindingDB,ChEMBL |
| Ki | 398.0 nM | PMID19338356 | BindingDB |
| Ki | 398.11 nM | PMID19338356 | ChEMBL |
| Ki | 522.0 nM | PMID24900484 | BindingDB,ChEMBL |
| Ki | 554.0 nM | PMID18083560 | PDSP,BindingDB,ChEMBL |
| Ki | 560.0 nM | PMID18363352, PMID20018510 | PDSP,BindingDB,ChEMBL |
| Ki | 697.0 nM | PMID18754613 | PDSP,BindingDB,ChEMBL |
| Ki | 790.0 nM | PMID18293908, PMID26756097, PMID19595596, PMID18579386, PMID20943290, PMID23085772, PMID21702498, PMID22548457, PMID20718492, PMID18680276 | PDSP,BindingDB,ChEMBL |
| Ki | 815.0 nM | PMID18448340 | BindingDB,ChEMBL |
| Ki | 851.14 nM | PMID21334892 | BindingDB,ChEMBL |
| Ki | 900.0 nM | PMID24900561, PMID22916707 | BindingDB,ChEMBL |
| Ki | 927.0 nM | PMID21428406, PMID19767206 | BindingDB,ChEMBL |
| Ki | 1000.0 nM | PMID19520572 | PDSP |
| Ki | 1400.0 nM | PMID27240274 | BindingDB,ChEMBL |
| Ki | 1569.0 nM | PMID19351113 | BindingDB,ChEMBL |
| Ki | 1580.0 nM | PMID18342403, PMID20047331, PMID14736243, PMID16140010, PMID15771428, PMID20363132 | PDSP,BindingDB,ChEMBL |
| Ki | 1640.0 nM | PMID17942307 | PDSP,BindingDB,ChEMBL |
| Ki | 1990.0 nM | PMID17979261 | PDSP,BindingDB,ChEMBL |
| Ki | 2511890.0 nM | PMID21183257 | ChEMBL |
| Kieq | 790.0 nM | PMID26756097 | ChEMBL |
| Log Ki | 2.85 nM | PMID10882356 | ChEMBL |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218