

You can:
| Name | Cannabinoid receptor 1 |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Cnr1 |
| Synonym | SKR6R Neuronal cannabinoid receptor Central cannabinoid receptor CB1R CB1 receptor [ Show all ] |
| Disease | N/A for non-human GPCRs |
| Length | 473 |
| Amino acid sequence | MKSILDGLADTTFRTITTDLLYVGSNDIQYEDIKGDMASKLGYFPQKFPLTSFRGSPFQEKMTAGDNSPLVPAGDTTNITEFYNKSLSSFKENEENIQCGENFMDMECFMILNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCRPSYHFIGSLAVADLLGSVIFVYSFVDFHVFHRKDSPNVFLFKLGGVTASFTASVGSLFLTAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCKKLQSVCSDIFPLIDETYLMFWIGVTSVLLLFIVYAYMYILWKAHSHAVRMIQRGTQKSIIIHTSEDGKVQVTRPDQARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMFPSCEGTAQPLDNSMGDSDCLHKHANNTASMHRAAESCIKSTVKIAKVTMSVSTDTSAEAL |
| UniProt | P20272 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL3571 |
| IUPHAR | 56 |
| DrugBank | N/A |
| Name | Rimonabant |
|---|---|
| Molecular formula | C22H21Cl3N4O |
| IUPAC name | 5-(4-chlorophenyl)-1-(2,4-dichlorophenyl)-4-methyl-N-piperidin-1-ylpyrazole-3-carboxamide |
| Molecular weight | 463.787 |
| Hydrogen bond acceptor | 3 |
| Hydrogen bond donor | 1 |
| XlogP | 6.5 |
| Synonyms | DSSTox_GSID_46453 HMS3657O15 LS-128155 4CH-015711 N-piperidino-5-(4-chlorophenyl)-1-(2,4-dichlorophenyl)-4-methyl-3-pyrazolecarboxamide [ Show all ] |
| Inchi Key | JZCPYUJPEARBJL-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C22H21Cl3N4O/c1-14-20(22(30)27-28-11-3-2-4-12-28)26-29(19-10-9-17(24)13-18(19)25)21(14)15-5-7-16(23)8-6-15/h5-10,13H,2-4,11-12H2,1H3,(H,27,30) |
| PubChem CID | 104850 |
| ChEMBL | CHEMBL111 |
| IUPHAR | 743 |
| BindingDB | 21278 |
| DrugBank | DB06155 |
Structure | 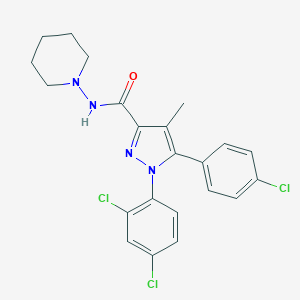 |
| Lipinski's druglikeness | This ligand has a partition coefficient log P greater than 5. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| Activity | -4.8 % | PMID17004712 | ChEMBL |
| Activity | 46.6 % | PMID17004712 | ChEMBL |
| EC50 | 0.17 nM | PMID18077343 | BindingDB,ChEMBL |
| EC50 | 56305.0 nM | PMID20845959 | BindingDB,ChEMBL |
| ED50 | 3.2 mg.kg-1 | PMID14736243 | ChEMBL |
| Emax | -37.8 % | PMID20845959 | ChEMBL |
| IC50 | 1.9 nM | PMID18243711 | BindingDB,ChEMBL |
| IC50 | 2.5 nM | PMID20584609 | BindingDB,ChEMBL |
| IC50 | 4.5 nM | PMID18337096 | BindingDB,ChEMBL |
| IC50 | 4.53 nM | PMID20045337, PMID20673729, PMID18954042 | BindingDB,ChEMBL |
| IC50 | 5.0 nM | PMID19596576, PMID19850473, PMID19328001, PMID19022666, PMID19269817 | BindingDB,ChEMBL |
| IC50 | 5.2 nM | PMID19128970 | BindingDB,ChEMBL |
| IC50 | 18.0 nM | PMID21741835 | BindingDB,ChEMBL |
| IC50 | 3920.0 nM | PMID17582659 | BindingDB,ChEMBL |
| Ki | 0.31 nM | PMID18448340 | BindingDB,ChEMBL |
| Ki | 0.65 nM | PMID19102698 | BindingDB,ChEMBL |
| Ki | 0.9 nM | PMID19683918 | BindingDB,ChEMBL |
| Ki | 1.18 nM | PMID12061874 | BindingDB,ChEMBL |
| Ki | 1.23 nM | PMID16451053 | BindingDB |
| Ki | 1.3 nM | PMID11906286 | BindingDB,ChEMBL |
| Ki | 1.8 nM | PMID12570386 | BindingDB,ChEMBL |
| Ki | 1.98 nM | PMID19128970 | BindingDB,ChEMBL |
| Ki | 4.0 nM | PMID15139773 | BindingDB |
| Ki | 6.0 nM | PMID19527048 | BindingDB,ChEMBL |
| Ki | 6.18 nM | PMID12061874 | BindingDB,ChEMBL |
| Ki | 6.5 nM | PMID19457667 | BindingDB,ChEMBL |
| Ki | 11.5 nM | PMID10052983 | BindingDB,ChEMBL |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218