

You can:
| Name | Muscarinic acetylcholine receptor M3 |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Chrm3 |
| Synonym | M3R M3 receptor HM4 Chrm-3 cholinergic receptor, muscarinic 3, cardiac [ Show all ] |
| Disease | N/A for non-human GPCRs |
| Length | 589 |
| Amino acid sequence | MTLHSNSTTSPLFPNISSSWVHSPSEAGLPLGTVTQLGSYNISQETGNFSSNDTSSDPLGGHTIWQVVFIAFLTGFLALVTIIGNILVIVAFKVNKQLKTVNNYFLLSLACADLIIGVISMNLFTTYIIMNRWALGNLACDLWLSIDYVASNASVMNLLVISFDRYFSITRPLTYRAKRTTKRAGVMIGLAWVISFVLWAPAILFWQYFVGKRTVPPGECFIQFLSEPTITFGTAIAAFYMPVTIMTILYWRIYKETEKRTKELAGLQASGTEAEAENFVHPTGSSRSCSSYELQQQGVKRSSRRKYGRCHFWFTTKSWKPSAEQMDQDHSSSDSWNNNDAAASLENSASSDEEDIGSETRAIYSIVLKLPGHSSILNSTKLPSSDNLQVSNEDLGTVDVERNAHKLQAQKSMGDGDNCQKDFTKLPIQLESAVDTGKTSDTNSSADKTTATLPLSFKEATLAKRFALKTRSQITKRKRMSLIKEKKAAQTLSAILLAFIITWTPYNIMVLVNTFCDSCIPKTYWNLGYWLCYINSTVNPVCYALCNKTFRTTFKTLLLCQCDKRKRRKQQYQQRQSVIFHKRVPEQAL |
| UniProt | P08483 |
| Protein Data Bank | 4daj, 4u14, 4u15, 4u16, 5zhp |
| GPCR-HGmod model | N/A |
| 3D structure model | This structure is from PDB ID 4daj. |
| BioLiP | BL0433837, BL0433838,BL0433839, BL0300355,BL0300356, BL0300353,BL0300354, BL0223908,BL0223909,BL0223910,, BL0300352 |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL320 |
| IUPHAR | 15 |
| DrugBank | N/A |
| Name | pirenzepine |
|---|---|
| Molecular formula | C19H21N5O2 |
| IUPAC name | 11-[2-(4-methylpiperazin-1-yl)acetyl]-5H-pyrido[2,3-b][1,4]benzodiazepin-6-one |
| Molecular weight | 351.41 |
| Hydrogen bond acceptor | 5 |
| Hydrogen bond donor | 1 |
| XlogP | 0.1 |
| Synonyms | IDI1_000127 L-S 519 NCGC00015836-02 11-[2-(4-methylpiperazin-1-yl)-acetyl]-5,11-dihydro-6H-pyrido[2,3-b][1,4]benzodiazepin-6-one NCGC00015836-10 [ Show all ] |
| Inchi Key | RMHMFHUVIITRHF-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C19H21N5O2/c1-22-9-11-23(12-10-22)13-17(25)24-16-7-3-2-5-14(16)19(26)21-15-6-4-8-20-18(15)24/h2-8H,9-13H2,1H3,(H,21,26) |
| PubChem CID | 4848 |
| ChEMBL | CHEMBL9967 |
| IUPHAR | 328 |
| BindingDB | 39341 |
| DrugBank | DB00670 |
Structure | 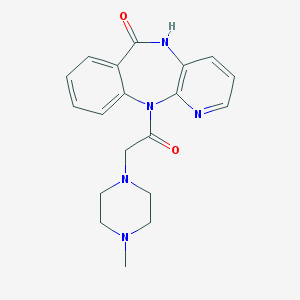 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| IC50 | 300.0 nM | PMID10571170 | BindingDB,ChEMBL |
| IC50 | 600.0 nM | PMID2754696 | BindingDB,ChEMBL |
| Ki | 28.0 nM | PMID8464039 | BindingDB,ChEMBL |
| Ki | 81.28 nM | PMID2250662 | BindingDB |
| Ki | 158.49 nM | PMID2188581 | BindingDB |
| Ki | 173.78 nM | PMID7932564, Bioorg. Med. Chem. Lett., (1995) 5:20:2325, PMID8246244, Bioorg. Med. Chem. Lett., (1995) 5:8:785 | ChEMBL |
| Ki | 173.789 nM | PMID8246244 | BindingDB |
| Ki | 174.0 nM | N/A | BindingDB |
| Ki | 199.526 nM | PMID1325587 | IUPHAR |
| Ki | 240.0 nM | PMID2066986 | BindingDB,ChEMBL |
| Ki | 320.0 nM | PMID11303063 | BindingDB |
| Ki | 450.0 nM | PMID2385234 | BindingDB |
| Ki | 720.0 nM | PMID11303063 | BindingDB |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218