| Synonyms | Merck dura Brand of Caffeine
3378-EP2371814A1
Dasin
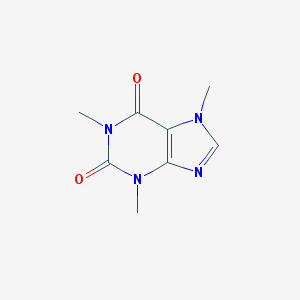
1,3,7-trimethyl-1,3,7-trihydropurine-2,6-dione
MLS001055341
58-08-2 solution, 20 mM in DMSO
DivK1c_000730
1,3,7-Trimethylpurine-2,6-dione
AB00051930_10
Eldiatric C
1-methyl-Theobromine
Caffeine solution
Alert-pep
FT-0664194
2,6-Dioxo-2,3,6,7-tetrahydro-1,3,7-trimethyl-1H-purine
Caffeine, 1mg/ml in methanol
AS-15340
HMS1920I09
3378-EP2269610A2
Caffeine, European Pharmacopoeia (EP) Reference Standard
BIM-0050216.0001
HSDB 36
3378-EP2295434A2
Caffeine, Sigma Reference Standard, vial of 250 mg
Bristol-Myers Squibb Brand of Caffeine
KBio1_000730
3378-EP2305696A2
CCG-38825
cafeine
Kofein
3378-EP2311801A1
Coffein [German]
caffeine (1,3,7-trimethylxanthine)
Lopac0_000228
3378-EP2314590A1
component of P-A-C Compound (Salt/Mix)
Propoxyphene Compound 65 (Salt/Mix)
RYYVLZVUVIJVGH-UHFFFAOYSA-N
Natural Caffeinum
SK-65 Compound
NCGC00015208-08
Spectrum_001301
NCGC00015208-17
STK177283
NCGC00090699-07
Theophylline, 7-methyl
NCI-C02733
Tri-Aqua
Nodoz Maximum Strength Caplets
Xanthine,3,7-trimethyl
P-A-C Analgesic Tablets
Percutafeine
Methylxanthine theophylline
DB00201
1,3,7-trimethyl-2,6-dioxo-1,2,3,6-tetrahydropurine
MolMap_000054
7-Methyltheophylline
DSSTox_RID_75448
1,3,7-Trimethylxanthine solution, 20 mM in DMSO
AC1Q3Z23
EU-0100228
1gfz
Caffeine solution, 20 mM in DMSO
Anhydrous caffeine
Guaranine
3,7-Dihydro-1,3,7-trimethyl-1H-purin-2,6-dion
Caffeine, anhydrous, 99%, FCC, FG
BDBM10849
HMS3260N17
3378-EP2277867A2
Caffeine, Pharmaceutical Secondary Standard; Certified Reference Material
Bio1_001451
I14-19223
3378-EP2305662A1
Caffeinum
C 0750
KBio2_006917
3378-EP2308562A2
CHEBI:27732
Caffedrine Caplets
Koffein [German]
3378-EP2311806A2
Coffeinum N
Caffeine 1.0 mg/ml in Methanol
Mateina
3378-EP2316458A1
CTK1G9501
Quick Pep
SCHEMBL5671
NCGC00015208-03
SPECTRUM1500155
NCGC00015208-12
SR-01000075187-4
NCGC00090699-02
Theine
NCGC00168808-01
Tirend
Nix Nap
Vivarin
NSC-755917
ZINC1084
PDSP1_001235
Phensal
3378-EP2316827A1
D0B3HD
07E4FB58-FD79-4175-8E3D-05BF96954522
Midol Maximum Strength
58-08-2
DHCplus
1,3,7-Trimethyl-3,7-dihydro-1H-purine-2,6-dione #
N1379
A.S.A. and Codeine Compound
EC 200-362-1
1,7-Trimethylxanthine
Caffeine melting point standard, United States Pharmacopeia (USP) Reference Standard
AI3-20154
FT-0623369
1l5q
Caffeine [BAN:JAN]
Anhydrous caffeine (TN)
Guaranine solution, 40 mM in DMSO
3,7-dihydro-1,3,7-trimethyl-1H-purine (9CI)
Caffeine, British Pharmacopoeia (BP) Reference Standard
BIDD:GT0632
HMS3715D13
3378-EP2289510A1
Caffeine, purum, anhydrous, >=99.0% (HPLC)
BRD-K02404261-001-03-5
InChI=1/C8H10N4O2/c1-10-4-9-6-5(10)7(13)12(3)8(14)11(6)2/h4H,1-3H
3378-EP2305682A1
Cafipel
Cafecon
KBioSS_001781
3378-EP2308870A2
CJ-00028
Caffeina [Italian]
Lanorinal
3378-EP2311842A2
component of Cafergot
CAFFEINE ANHYDROUS
MEGxp0_001350
Probes2_000128
Republic Drug Brand of Caffeine
Seid Brand of Caffeine
NCGC00015208-06
Spectrum4_001782
NCGC00015208-15
ST057528
NCGC00090699-05
Theobromine, 1-methyl-
NCGC00259234-01
Tox21_300010
Nodaca
WLN: T56 BN DN FNVNVJ B1 F1 H1
O926
Pep-Back
Methyltheobromide
3G6A5W338E
DB-023002
1,3,7-trimethyl-1H-purine-2,6(3H,7H)-dione
MLS001056714
58-08-2 solution, 40 mM in DMSO
DSSTox_CID_232
1,3,7-Trimethylxanthine
AC-12774
Enerjets
1-methyltheobromine
Caffeine solution, 1.0 mg/mL in methanol, ampule of 1 mL, certified reference material
AN-23595
GlaxoSmithKline Brand of Caffeine
2a3b
Caffeine, 98.5%, specified according to the req. of USP/BP
Bayer Select Headache Pain (Salt/Mix)
HMS2091O11
3378-EP2275420A1
Caffeine, meets USP testing specifications, anhydrous
Bio1_000473
Hycomine
3378-EP2301937A1
Caffeine, synthetic
BRN 0017705
KBio2_001781
3378-EP2305697A2
CCRIS 1314
Cafergot (Salt/Mix)
Kofein [Czech]
3378-EP2311802A1
Coffeine
Caffeine (natural)
LP00228
3378-EP2314593A1
component of Percodan (Salt/Mix)
Propoxyphene Compound-65
SBB006474
NCGC00015208-01
SMR000326667
NCGC00015208-10
SR-01000075187
NCGC00015208-18
teina
NCGC00090699-08
Thompson Brand 1 of Caffeine
NCIOpen2_008255
Ultra Pep-Back
NSC 5036
Z112207564
Passauer Brand of Caffeine
peyona
CU-01000012617-3
Mettler-Toledo Calibration substance ME 18872, Caffeine, analytical standard, for the calibration of the thermosystem 900, traceable to primary standards (LGC)
5-26-13-00558 (Beilstein Handbook Reference)
Dexitac
1,3,7-Trimethyl-2,6-dioxopurine
Monohydrate Caffeine
71701-02-5
DTXSID0020232
1,3,7-Trimethylxanthine solution, 40 mM in DMSO
Caffeine hydrous
ACN-034787
F3371-0262
1H-Purine-2, 3,7-dihydro-1,3,7-trimethyl-
Caffeine solution, 40 mM in DMSO
Anhydrous caffeine (JP15)
Guaranine solution, 10 mM in DMSO
3,7-Dihydro-1,3,7-trimethyl-1H-purin-2,6-dion (coffein)
Caffeine, anhydrous, tested according to Ph.Eur.
Berlin-Chemie Brand of Caffeine
HMS3372J18
3378-EP2280003A2
Caffeine, PharmaGrade, EP, Manufactured under appropriate GMP controls for pharma or biopharmaceutical production
bmse000206
I14-4386
3378-EP2305671A1
caffenium
C07481
KBio3_001141
3378-EP2308840A1
Caffein
KS-0000478B
3378-EP2311808A1
Coffeinum Purrum
Caffeine 10 microg/mL in Methanol
Maximum Strength Snapback Stimulant Powders
3378-EP2316825A1
Pierre Fabre Brand of Caffeine
Quick-Pep
SDCCGMLS-0064595.P001
NCGC00015208-04
Spectrum2_001261
NCGC00015208-13
SR-01000075187-7
NCGC00090699-03
Theine, methyltheobromine
NCGC00168808-02
TNP00310
No Doz
Wake-Up
NSC5036
PDSP2_001000
3378-EP2316828A1
Darvon compound-65
1, 3, 7-Trimethylxanthine
Miudol
58-08-2 solution, 10 mM in DMSO
Diurex
1,3,7-trimethyl-3,7-dihydropurine-2,6-dione
AB00051930-09
EINECS 200-362-1
1-3-7-TRIMETHYLXANTHINE
Caffeine Pure
AKOS000121334
FT-0656253
1l7x
Caffeine [USP:BAN:JAN]
ARONIS25359
H2815
3,7-Dihydro-1,3,7-trimethyl-1H-purine-2,6-dione
Caffeine, certified reference material, TraceCERT(R)
BIDD:PXR0172
HMS502E12
3378-EP2295409A1
Caffeine, SAJ special grade, >=98.5%
BRD-K02404261-001-07-6
KB-48627
3378-EP2305695A2
CAS-58-08-2
Cafeina
Keep Alert
3378-EP2308879A1
Coffein
Lopac-C-0750
3378-EP2314588A1
component of Dilone (Salt/Mix)
Caffeine for system suitability, European Pharmacopoeia (EP) Reference Standard
Melting point standard 235-237C, analytical standard
Propoxyphene Compound 65
Respia (TN)
N2379
SK 65 Compound
NCGC00015208-07
Spectrum5_000423
NCGC00015208-16
Stim
NCGC00090699-06
Theophylline Me
NCGC00260913-01
Tox21_500228
NoDoz Caplets and Chewable Tablets
Xanthine, 1,3,7-trimethyl
Organex
Percoffedrinol N
Methyltheobromine
3g6m
1,3,7-trimethyl-2,3,6,7-tetrahydro-1H-purine-2,6-dione
MLS001066409
7-methyl Theophylline
DSSTox_GSID_20232
1,3,7-Trimethylxanthine solution, 10 mM in DMSO
AC1L1DV2
Ercatab
Caffeine solution, 10 mM in DMSO
Anacin Maximum Strength
GTPL407
3,3,7-trimethyl-1H-purine-2,6-dione
Caffeine, anhydrous
BBL016491
HMS2232M13
3378-EP2277848A1
Caffeine, Monohydrate
Bio1_000962
Hycomine Compound
3378-EP2305219A1
Caffeine, United States Pharmacopeia (USP) Reference Standard
BSPBio_001921
KBio2_004349
3378-EP2305698A2
CFF
Caffedrine
Koffein
3378-EP2311803A1
Coffeinum
Caffeine (USP)
LS-237
3378-EP2316457A1
CS-M0795
PubChem9436
SBI-0050216.P004
NCGC00015208-02
SPBio_001222
NCGC00015208-11
SR-01000075187-1
NCGC00090699-01
Thein
NCGC00090699-09
Thompson Brand 2 of Caffeine
NINDS_000730
UNII-3G6A5W338E
NSC-5036
ZB000243
PDSP1_001016
Pharmakon1600-01500155
3378-EP2316826A1
D00528
MFCD00005758
5-26-13-00558 (Beilstein)
Dexitac Stay Alert Stimulant
1,3,7-Trimethyl-3,7-dihydro-1H-purine-2,6-dione
Monomethyl derivative of Theophylline
95789-13-2
Durvitan
1,7-Trimethyl-2,6-dioxopurine
Caffeine Melting Point Standard, Pharmaceutical Secondary Standard; Certified Reference Material
ACon1_000085
FEMA No. 2224
1H-Purine-2,6-dione, 3,7-dihydro-1,3,7-trimethyl-
Caffeine solution, analytical standard, 1.0 mg/mL in methanol
Anhydrous caffeine (JP17)
Guaranine solution, 20 mM in DMSO
3,7-dihydro-1,3,7-trimethyl-1H-purine
Caffeine, BioXtra
BIDD:ER0554
HMS3435F10
3378-EP2284160A1
Caffeine, powder, ReagentPlus(R)
BRD-K02404261-001-02-7
IDI1_000730
3378-EP2305677A1
Caffine
Cafamil
KBioGR_002325
3378-EP2308867A2
CHEMBL113
Caffeina
L000155
3378-EP2311829A1
component of A.S.A. Compound (Salt/Mix)
Caffeine 99%
MCULE-3362813910
Probes1_000150
Refresh'n
SDCCGMLS-0064595.P002
NCGC00015208-05
Spectrum3_000321
NCGC00015208-14
SR-01000075187-8
NCGC00090699-04
Theobromine Me
NCGC00254057-01
Tox21_201685
No-Doz
Wigraine
NSC755917
PDSP2_001219 [ Show all ] |
|---|


![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218