

You can:
| Name | Mu-type opioid receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | OPRM1 |
| Synonym | hMOP M-OR-1 MOP opioid receptor, mu 1 opioid receptor [ Show all ] |
| Disease | Diarrhea Inflammatory disease Pain Major depressive disorder Migraine [ Show all ] |
| Length | 400 |
| Amino acid sequence | MDSSAAPTNASNCTDALAYSSCSPAPSPGSWVNLSHLDGNLSDPCGPNRTDLGGRDSLCPPTGSPSMITAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALATSTLPFQSVNYLMGTWPFGTILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIINVCNWILSSAIGLPVMFMATTKYRQGSIDCTLTFSHPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGSKEKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALVTIPETTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCFREFCIPTSSNIEQQNSTRIRQNTRDHPSTANTVDRTNHQLENLEAETAPLP |
| UniProt | P35372 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | P35372 |
| 3D structure model | This predicted structure model is from GPCR-EXP P35372. |
| BioLiP | N/A |
| Therapeutic Target Database | T47768 |
| ChEMBL | CHEMBL233 |
| IUPHAR | 319 |
| DrugBank | BE0000770 |
| Name | naloxone |
|---|---|
| Molecular formula | C19H21NO4 |
| IUPAC name | (4R,4aS,7aR,12bS)-4a,9-dihydroxy-3-prop-2-enyl-2,4,5,6,7a,13-hexahydro-1H-4,12-methanobenzofuro[3,2-e]isoquinolin-7-one |
| Molecular weight | 327.38 |
| Hydrogen bond acceptor | 5 |
| Hydrogen bond donor | 2 |
| XlogP | 2.1 |
| Synonyms | DSSTox_RID_76986 HSDB 3279 MLS000069540 (4R,4aS,7aR,12bS)-4a,9-dihydroxy-3-prop-2-enyl-2,4,5,6,7a,13-hexahydro-1H-4,12-methanobenzofuro[3,2-e]isoquinoline-7-one;hydrochloride Morphinan-6-one, 4,5-epoxy-3,14-dihydroxy-17-(2-propenyl)-, (5alpha)-(9CI) [ Show all ] |
| Inchi Key | UZHSEJADLWPNLE-GRGSLBFTSA-N |
| Inchi ID | InChI=1S/C19H21NO4/c1-2-8-20-9-7-18-15-11-3-4-12(21)16(15)24-17(18)13(22)5-6-19(18,23)14(20)10-11/h2-4,14,17,21,23H,1,5-10H2/t14-,17+,18+,19-/m1/s1 |
| PubChem CID | 5284596 |
| ChEMBL | CHEMBL80 |
| IUPHAR | 1676, 1638 |
| BindingDB | 50000788, 54795 |
| DrugBank | DB01183 |
Structure | 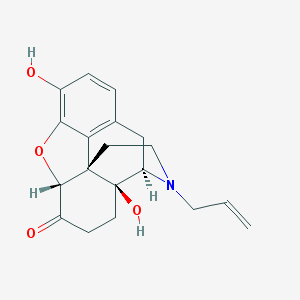 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| N/A | N/A | DrugBank | |
| Activity | 0.1 % | PMID18207400, PMID17266203 | ChEMBL |
| Activity | 50.0 % | PMID19836950 | ChEMBL |
| Activity | 90.0 % | PMID8496700 | ChEMBL |
| Activity | 97.0 % | PMID26035635, PMID21667972 | ChEMBL |
| Emax | 13.0 % | PMID17407276 | ChEMBL |
| IC50 | 1.87 nM | PMID24171469 | ChEMBL |
| IC50 | 2.0 nM | PMID11585443 | BindingDB,ChEMBL |
| IC50 | 7.3 nM | PMID18313920, PMID14643346, PMID17149859, PMID17149858 | BindingDB,ChEMBL |
| IC50 | 20.0 nM | DrugMatrix in vitro pharmacology data | ChEMBL |
| IC50 | 23.0 nM | PMID17407276 | BindingDB,ChEMBL |
| Imax | 92.0 % | PMID17407276 | ChEMBL |
| Inhibition | 100.1 % | PMID23659286 | ChEMBL |
| Kd | 0.2188 nM | PMID18207400, PMID17266203 | ChEMBL |
| Ke | 2.3 nM | PMID21570305, PMID21684752, PMID22341895, PMID25599950, PMID21247164, PMID23618710, PMID19053757, PMID17625813, PMID20055417 | ChEMBL |
| Ki | 0.23 nM | PMID21482470 | ChEMBL |
| Ki | 0.23 nM | PMID21482470 | BindingDB |
| Ki | 0.5623 nM | PMID19527931 | ChEMBL |
| Ki | 0.562341 nM | PMID19527931 | BindingDB |
| Ki | 0.57 nM | PMID21621410 | ChEMBL |
| Ki | 0.57 nM | PMID21621410 | BindingDB |
| Ki | 0.66 nM | PMID19027293 | ChEMBL |
| Ki | 0.66 nM | PMID19027293 | BindingDB |
| Ki | 0.79 nM | PMID17407276 | ChEMBL |
| Ki | 0.79 nM | PMID17407276 | BindingDB |
| Ki | 0.93 nM | PMID8114680 | BindingDB |
| Ki | 0.98 nM | PMID20055417 | ChEMBL |
| Ki | 0.98 nM | PMID20055417 | BindingDB |
| Ki | 1.1 nM | PMID26632862 | BindingDB |
| Ki | 1.148 nM | PMID26632862 | ChEMBL |
| Ki | 1.25893 nM | PMID9686407 | IUPHAR |
| Ki | 1.259 nM | PMID26632862 | ChEMBL |
| Ki | 1.3 nM | PMID26632862 | BindingDB |
| Ki | 1.35 nM | PMID7932177 | BindingDB |
| Ki | 1.4 nM | PMID7815359, PMID9686407 | BindingDB |
| Ki | 1.5 nM | PMID19836950 | BindingDB,ChEMBL |
| Ki | 1.6 nM | PMID8496700 | BindingDB,ChEMBL |
| Ki | 2.27 nM | PMID21621410 | ChEMBL |
| Ki | 2.3 nM | MedChemComm, (2016) 7:2:317, PMID25062506, PMID21621410 | BindingDB,ChEMBL |
| Ki | 3.6 nM | PMID12747782 | BindingDB |
| Ki | 3.63 nM | PMID12747782 | ChEMBL |
| Ki | 3.7 nM | PMID18313920, PMID14643346, PMID17149859, PMID17149858 | BindingDB,ChEMBL |
| Ki | 4.2 nM | PMID21621410 | BindingDB |
| Ki | 4.23 nM | PMID21621410 | ChEMBL |
| Ki | 8.063 nM | DrugMatrix in vitro pharmacology data | ChEMBL |
| Ki | 354.0 nM | PMID25268943 | BindingDB,ChEMBL |
| Ki | 1000.0 nM | PMID7932177, PMID7815359 | BindingDB |
| pKb | 9.09 - | PMID19527931 | ChEMBL |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218