

You can:
| Name | Kappa-type opioid receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | OPRK1 |
| Synonym | K-OR-1 KOPr OP2 KOP KOR-1 [ Show all ] |
| Disease | Obesity Opiate dependence Inflammatory bowel disease Erythema Diarrhea-predominant IBS [ Show all ] |
| Length | 380 |
| Amino acid sequence | MDSPIQIFRGEPGPTCAPSACLPPNSSAWFPGWAEPDSNGSAGSEDAQLEPAHISPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVISIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFPDDDYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGSREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSTSHSTAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFPLKMRMERQSTSRVRNTVQDPAYLRDIDGMNKPV |
| UniProt | P41145 |
| Protein Data Bank | 6b73, 4djh |
| GPCR-HGmod model | P41145 |
| 3D structure model | This structure is from PDB ID 6b73. |
| BioLiP | BL0402244,BL0402246, BL0224693,BL0224694, BL0402243,BL0402245 |
| Therapeutic Target Database | T60693 |
| ChEMBL | CHEMBL237 |
| IUPHAR | 318 |
| DrugBank | BE0000632 |
| Name | U50488 |
|---|---|
| Molecular formula | C19H26Cl2N2O |
| IUPAC name | 2-(3,4-dichlorophenyl)-N-methyl-N-[(1R,2R)-2-pyrrolidin-1-ylcyclohexyl]acetamide |
| Molecular weight | 369.33 |
| Hydrogen bond acceptor | 2 |
| Hydrogen bond donor | 0 |
| XlogP | 4.4 |
| Synonyms | 3,4-Dichloro-N-methyl-N-(2-(1-pyrrolidinyl)-cyclohexyl)-benzeneacetamide, (trans)-Isomer Benzeneacetamide, 3,4-dichloro-N-methyl-N-(2-(1-pyrrolidinyl)cyclohexyl)-, trans- Lopac-U-110 SCHEMBL332193 U-50488H [ Show all ] |
| Inchi Key | VQLPLYSROCPWFF-QZTJIDSGSA-N |
| Inchi ID | InChI=1S/C19H26Cl2N2O/c1-22(19(24)13-14-8-9-15(20)16(21)12-14)17-6-2-3-7-18(17)23-10-4-5-11-23/h8-9,12,17-18H,2-7,10-11,13H2,1H3/t17-,18-/m1/s1 |
| PubChem CID | 3036289 |
| ChEMBL | CHEMBL441765 |
| IUPHAR | 1652 |
| BindingDB | 50000296 |
| DrugBank | N/A |
Structure | 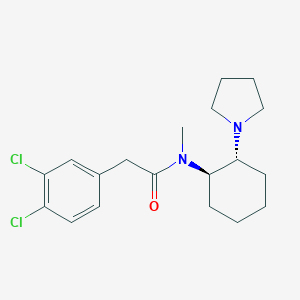 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| Activity | 19.0 % | PMID19640720 | ChEMBL |
| Activity | 85.0 - | PMID7515442 | ChEMBL |
| EC50 | 1.4 nM | PMID17981041 | BindingDB,ChEMBL |
| EC50 | 2.9 nM | PMID19147366 | BindingDB,ChEMBL |
| EC50 | 3.4 nM | PMID16051487, PMID16945525 | BindingDB,ChEMBL |
| EC50 | 4.2 nM | PMID22439881 | BindingDB,ChEMBL |
| EC50 | 4.5 nM | PMID16777411, PMID15993589, PMID15869877 | BindingDB,ChEMBL |
| EC50 | 5.7 nM | PMID22204910 | BindingDB,ChEMBL |
| EC50 | 6.3 nM | PMID20801035 | BindingDB,ChEMBL |
| EC50 | 9.3 nM | PMID23134120 | BindingDB |
| EC50 | 9.31 nM | PMID23134120 | ChEMBL |
| EC50 | 11.0 nM | PMID26330078 | BindingDB |
| EC50 | 14.0 nM | PMID10893307 | BindingDB |
| EC50 | 14.4 nM | PMID10893307 | ChEMBL |
| EC50 | 30.0 nM | PMID18380425 | BindingDB,ChEMBL |
| EC50 | 36.0 nM | PMID17935988, PMID19282177, PMID23434225, PMID16942039, PMID19027293, PMID19091564, PMID18417347, PMID23142613 | BindingDB,ChEMBL |
| EC50 | 46.0 nM | PMID17407276, PMID17276685, PMID14613319, PMID17433695 | BindingDB,ChEMBL |
| EC50 | 630.96 nM | PMID27234885 | ChEMBL |
| EC50 | 631.0 nM | PMID27234885 | BindingDB |
| ED50 | 27.0 nM | PMID25599950 | ChEMBL |
| Efficacy | 100.0 % | PMID16051487, PMID16777411, PMID15869877 | ChEMBL |
| Emax | 77.0 % | PMID17935988, PMID19282177, PMID19027293, PMID16942039, PMID23434225, PMID19091564, PMID18417347, PMID23142613 | ChEMBL |
| Emax | 93.0 % | PMID23134120 | ChEMBL |
| Emax | 99.0 % | PMID25599950 | ChEMBL |
| Emax | 100.0 % | PMID19147366 | ChEMBL |
| Emax | 110.0 % | PMID17276685, PMID17407276, PMID14613319, PMID17433695 | ChEMBL |
| Emax | 140.0 % | PMID22439881 | ChEMBL |
| IC50 | 0.3 nM | PMID25087049 | ChEMBL |
| IC50 | 0.3 nM | PMID25087049 | BindingDB |
| IC50 | 0.55 nM | PMID18588282 | ChEMBL |
| IC50 | 0.64 nM | PMID27876250 | ChEMBL |
| IC50 | 0.69 nM | PMID11906279 | BindingDB |
| IC50 | 0.69 nM | PMID11906279 | ChEMBL |
| IC50 | 0.75 nM | PMID23466604 | ChEMBL |
| IC50 | 0.84 nM | PMID23582449 | ChEMBL |
| IC50 | 1.99 nM | PMID19640720 | BindingDB,ChEMBL |
| IC50 | 2.5 nM | PMID18983139 | ChEMBL |
| IC50 | 6.4 nM | PMID27876250 | BindingDB |
| IC50 | 370.0 nM | PMID8393489, PMID1310743 | BindingDB,ChEMBL |
| Ki | 0.199526 - 15.8489 nM | PMID9686407, PMID7869844, PMID7624359, PMID9262330, PMID6129321, PMID14718611 | IUPHAR |
| Ki | 0.2 nM | PMID23134120 | ChEMBL |
| Ki | 0.2 nM | PMID23134120 | BindingDB |
| Ki | 0.5 nM | PMID23466604 | ChEMBL |
| Ki | 0.69 nM | PMID20478711 | BindingDB,ChEMBL |
| Ki | 0.73 nM | PMID21621410 | BindingDB |
| Ki | 0.95 nM | PMID21621410 | BindingDB |
| Ki | 1.2 nM | PMID20801035 | BindingDB,ChEMBL |
| Ki | 1.4 nM | PMID16051487, PMID16945525, PMID16777411, PMID15869877, PMID15993589 | BindingDB,ChEMBL |
| Ki | 1.5 nM | PMID20441176 | BindingDB,ChEMBL |
| Ki | 1.6 nM | PMID19147366 | BindingDB,ChEMBL |
| Ki | 1.7 nM | PMID18983139 | ChEMBL |
| Ki | 2.2 nM | PMID17981041 | BindingDB,ChEMBL |
| Ki | 2.7 nM | PMID26330078 | BindingDB |
| Ki | 2.9 nM | PMID22995061, PMID25051243 | BindingDB,ChEMBL |
| Ki | 4.5 nM | PMID22204910 | BindingDB,ChEMBL |
| Ki | 6.08 nM | PMID27185012 | ChEMBL |
| Ki | 6.1 nM | PMID27185012 | BindingDB |
| Ki | 15.0 nM | Bioorg. Med. Chem. Lett., (1992) 2:7:715, | BindingDB,ChEMBL |
| Ratio | 28.0 - | PMID9822548 | ChEMBL |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218