| Synonyms | DSSTox_GSID_21327
INT-0010
LS-975
QCD 84924
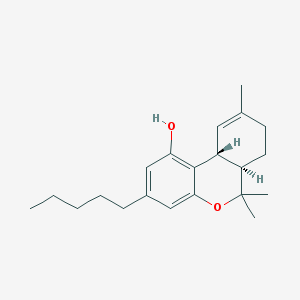
Tetrahydro-6,6,9-trimethyl-3-pentyl-6H-dibenzo(b,d)pyran-1-ol
Tox21_112616
US9416103, Delta9-THC
Delta9-Tetrahydrocannabinol solution, 1.0 mg/mL in methanol, analytical standard, for drug analysis
Dronabinol (USP/INN)
3ls4
(-)-delta(sup 1)-3,4-trans-Tetrahydrocannabinol
6H-Dibenzo[b, 6a,7,8,10a-tetrahydro-6,6,9-trimethyl-3-pentyl-
(-)-Delta9-THC (Dronabinol) 1.0 mg/ml in Methanol
9-delta-Tetrahydrocannabinol
(10R,10aR)-6,6,9-Trimethyl-3-pentyl-6a,7,8,10a-tetrahydro-6H-benzo[c]chromen-1-ol
BDBM50007391
(6aS,10aS)-6,6,9-trimethyl-3-pentyl-6a,7,8,10a-tetrahydro-6H-benzo[c]chromen-1-ol
Cannabinol, Delta1-tetrahydro- (7CI)
.DELTA.1-THC
CHEBI:66964
1-trans-delta-9-Tetrahydrocannabinol
D0P1FO
14146-43-1
delta(9)-Tetrahydrocannabinol
23623-EP2298772A1
delta-9 Tetrahydrocannabinol
Epitope ID:224552
L-.delta.1-trans-Tetrahydrocannabinol
Namisol
SCHEMBL4609
Tetrahydrocannabinol delta9
trans-delta-9-Tetrahydrocannabinol
delta1-Tetrahydrocannabinol (VAN)
delta9-trans-Tetrahydrocannabinol
Dronabinol [USAN:INN]
6,6,9-Trimethyl-3-pentyl-6a,7,8,10a-tetrahydro-6H-benzo[c]chromen-1-ol(delta9-THC(delta9-tetrahydrocannabinol))
(-)-.DELTA.1-Tetrahydrocannabinol
6H-Dibenzo
(-)-delta9-(trans)-Tetrahydrocannabinol
6H-dibenzo[b,d]pyran-1-ol, 6a,7,8,10a-tetrahydro-6,6,9-trimethyl-3-pentyl-, (6aR,10aR)-
(-)-trans-.DELTA.9-Tetrahydrocannabinol
9-THC
(6AR,10AR)-6,6,9-TRIMETHYL-3-PENTYL-6H,6AH,7H,8H,10AH-BENZO[C]ISOCHROMEN-1-OL
C06972
(l)-delta1-Tetrahydrocannabinol
Cannabinol, tetrahydro-(6CI)
.DELTA.9-trans-Tetrahydrocannabinol
Compassia
124699-EP2289509A2
DEA No. 7369
1972-08-3
delta(sup 1)-Tetrahydrocannabinol
23623-EP2314580A1
delta-9-THC
Hashish
L-trans-.delta.9-Tetrahydrocannabinol
NSC134454
Synthetic THC in sesame oil / soft gelatin
THC
UNII-7J8897W37S
delta9-Tetrahydrocannabinol (VAN)
Drona binol
Dronabinolum [Latin]
6,6,9-Trimethyl-3-pentyl-7,8,9,10-tetrahydro-6H-dibenzo(b,d)pyran-1-ol
(-)-.DELTA.9-trans-Tetrahydrocannabinol
6H-Dibenzo(b,d)pyran-1-ol, 6a,7,8,10a-tetrahydro-6,6,9-trimethyl-3-pentyl-, (6aR-trans)-
(-)-Delta9-Tetrahydrocannabinol solution, ~1 mg/mL in ethanol, analytical standard, for drug analysis
7J8897W37S
(-)-trans-Delta9-THC
AC1Q2VRZ
(6aR-trans)-6a,7,8,10a-Tetrahydro-6,6,9-trimethyl-3-pentyl-6H-dibenzo(b,d)pyran-1-ol
Cannabinol, 1-trans-delta(sup 9)-tetrahydro-
.delta.-9-THC
CAT-310
1-trans-delta(sup 9)-Tetrahydrocannabinol
D00306
1363-19-5
delta(1)-Tetrahydrocannabinol
23623-EP2298764A1
delta(sup 9)-Thc
3-Pentyl-6,6,9-trimethyl-6a,7,8,10a-tetrahydro-6H-dibenzo(b,d)pyran-1-ol
DSSTox_RID_76083
INT-0010/06
Marinol
QCD-84924
Tetrahydro-6,6,9-trimethyl-3-pentyl-6H-dibenzo[b,d]pyran-1-ol
trans-.DELTA.9-Tetrahydrocannabinol
ZINC1530625
delta-THC
Delta9-Tetrahydrocannabinol solution, ethanol solution
Dronabinol in sesame oil and encapsulated in a soft gelatin capsule in a U.S. FDA approved drug product
5957-27-7
(-)-(6aR,10aR)-6,6,9-trimethyl-3-pentyl-6a,7,8,10a-tetrahydro-6H-benzo[c]chromen-1-ol
6465-30-1
(-)-delta(sup 1)-3,4-trans-Tetrahydrocannabinol(l)-delta(sup 1)-Tetrahydrocannabinol
6H-Dibenzo[b, 6a,7,8,10a-tetrahydro-6,6,9-trimethyl-3-pentyl-, (6aR-trans)-
(-)-Delta9-THC (Dronabinol) 5.0 mg/ml in Methanol
9-ene-Tetrahydrocannabinol
(6aR,10aR)-6,6,9-trimethyl-3-pentyl-6a,7,8,10a-tetrahydro-6H-benzo[c]chromen-1-ol
BDBM60994
(L)-.delta.1-Tetrahydrocannabinol
Cannabinol, Delta1-tetrahydro-(7CI)
.DELTA.9-Tetrahydrocannabinol
1-trans-delta9-Tetrahydrocannabinol
148820-EP2289509A2
delta(9)-Tetrahydrocannibinol
23623-EP2308839A1
delta-9-tetrahydrocannabinol
Ganja
L-delta(sup 1)-tetrahydrocannabinol
NSC 134454
SP 104
Tetrahydrocannabinols (-)-delta1-3,4-trans-form
trans-delta9-Tetrahydrocannabinol
delta1-THC
Deltanyne
Dronabinol [USAN:USP:INN]
6,6,9-Trimethyl-3-pentyl-6a,7,8,10a-tetrahydro-6H-benzo[c]chromen-1-ol(DeltaE-9-THC)
(-)-.DELTA.9-Tetrahydrocannabinol
6H-Dibenzo(b,d)pyran-1-ol, 6a,7,8,10a-tetrahydro-3-pentyl-6,6,9-trimethyl- (6ar-trans)-
(-)-delta9-Tetrahydrocannabinol
6H-Dibenzo[b,d]pyran-1-ol, 6a,7,8,10a-tetrahydro-6,6,9-trimethyl-3-pentyl-, (6aR-trans)-
(-)-trans-Delta1-Tetrahydrocannabinol
Abbott 40566
(6aR,10aR)-6a,7,8,10a-Tetrahydro-6,6,9-trimethyl-3-pentyl-6H-dibenzo(b,d)pyran-1-ol
Cannabinol, .DELTA.1-tetrahydro-
(S)-6,6,9-Trimethyl-3-pentyl-6a,7,8,10a-tetrahydro-6H-benzo[c]chromen-1-ol
Cannabis resin
1-trans-.delta.(sup9)-tetrahydrocannabinol
CTK8D8346
124699-EP2295426A1
DEA No. 7370
23623-EP2269989A1
delta(sup 1)-Thc
DSSTox_CID_1327
HSDB 6471
L-trans-delta9-Tetrahydrocannabinol
PDSP2_000714
Tetrahydro-6,6,9-trimethyl-3-pentyl-6H-di benzo[b,d]pyran-1-ol
THC-delta-9
UNII-K4H93P747O component CYQFCXCEBYINGO-IAGOWNOFSA-N
Delta9-Tetrahydrocannabinol solution
6,9-Trimethyl-3-pentyl-7,8,9,10-tetrahydro-6H-dibenzo[b,d]pyran-1-ol
(-)-3,4-trans-Delta1-Tetrahydrocannabinol
6H-Dibenzo(b,d)pyran-1-ol, 6a,7,8,10a-tetrahydro-6,6,9-trimethyl-3-pentyl-, trans-
(-)-Delta9-THC (Dronabinol) 0.1 mg/ml in Methanol
8,8-Dimethyl-11-methylene-5-pentyl-3,4,8a,9,10,11,12,12a-octahydro-2H,8H-1,7-dioxa-benzo[c]phenanthrene
(-)-trans-Delta9-THC solution, 1.0 mg/mL in methanol, ampule of 1 mL, certified reference material
API0002497
(6aR-trans)-6a,7,8,10a-Tetrahydro-6,6,9-trimethyl-3-pentyl-6H-dibenzo[b,d]pyran-1-ol
Cannabinol, delta1-tetrahydro-
.DELTA.1-Tetrahydrocannabinol
CCRIS 4726
1-trans-delta(sup9)-tetrahydrocannabinol
D0L4AI
14146-29-3
delta(1)-THC
23623-EP2298765A1
delta(sup9)-Thc
3-pentyl-6,6,9-trimethyl-6a,7,8,10a-tetrahydro-6H-dibenzo[b,d]pyran-1-ol
DTXSID6021327
L-.delta.1-Tetrahydrocannabinol
Marinol (TN)
Relivar
TETRAHYDROCANNABINOL
trans-6a,7,8,10a-Tetrahydro-6,6,9-trimethyl-3-pentyl-6H-dibenzo(b,d)pyran-1-ol
delta1-Tetrahydrocannabinol
delta9-THC
Dronabinol in sesame oil in soft gelatin capsule
6,6,9-Trimethyl-3-pentyl-6a,7,8,10a-tetrahydro-6H-benzo[c]chromen-1-ol
(-)-.delta.(sup9)-trans-Tetrahydrocannabinol
6H-Dibe nzo[b,d]pyran-1-ol,6a,7,8,10a-tetrahydro-6,6,9-trimethyl-3-pentyl-, (6aR-trans)-
(-)-delta1-Tetrahydrocannabinol
6H-Dibenzo[b, 6a,7,8,10a-tetrahydro-6,6,9-trimethyl-3-pentyl-, trans-
(-)-delta9-trans-Tetrahydrocannabinol
9-tetrahydrocannabinol
(6aR,10aR)-6,6,9-trimethyl-3-pentyl-6a,7,8,10a-tetrahydrobenzo[c]chromen-1-ol
BIDD:GT0427
(l)-delta(sup 1)-Tetrahydrocannabinol
Cannabinol, tetrahydro- (6CI)
.DELTA.9-THC
CHEMBL465
124699-EP2272825A2
DB00470
14C-.DELTA.1-Tetrahydrocannabinol
delta(9)-THC
23623-EP2314298A1
Delta-9-Tetrahydrocannabinol, United States Pharmacopeia (USP) Reference Standard
GTPL2424
L-delta1-trans-Tetrahydrocannabinol
NSC-134454
Syndros
Tetranabinex
trans-tetrahydrocannabinol
delta9-Tetrahydrocannabinol
DRG-0138
Dronabinolum
6,6,9-trimethyl-3-pentyl-6a,7,8,10a-tetrahydro-6H-dibenzo[b,d]pyran-1-ol
(-)-.DELTA.9-THC
6H-Dibenzo(b,d)pyran-1-ol, 6a,7,8,10a-tetrahydro-6,6,9-trimethyl-3-pentyl-, (6aR,10aR)-
(-)-Delta9-Tetrahydrocannabinol solution
6H-Dibenzo[b,d]pyran-1-ol,6a,7,8,10a-tetrahydro-6,6,9-trimethyl-3-pentyl-, (6aR-trans)-
(-)-trans-delta9-Tetrahydrocannabinol
AC1L277U
(6aR,10aR)-6a,7,8,10a-Tetrahydro-6,6,9-trimethyl-3-pentyl-6H-dibenzo[b,d]pyran-1-ol
Cannabinol, 1-trans-.delta.(sup9)-tetrahydro-
.delta.(sup9)-THC
CAS-1972-08-3
1-trans-D9-Tetrahydrocannabinol
CYQFCXCEBYINGO-IAGOWNOFSA-N
124699-EP2295427A1
delta 9-Tetrahydrocannabinol
23623-EP2275420A1
delta(sup 9)-Tetrahydrocannabinol
26108-45-2 [ Show all ] |
|---|


![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218