

You can:
| Name | Cannabinoid receptor 1 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | CNR1 |
| Synonym | CB1 Central cannabinoid receptor SKR6R THC receptor CB1R [ Show all ] |
| Disease | Obesity; Diabetes Chemotherapy-induced nausea Diabetes; Obesity Drug abuse Hypertension; Diabetes; Obesity [ Show all ] |
| Length | 472 |
| Amino acid sequence | MKSILDGLADTTFRTITTDLLYVGSNDIQYEDIKGDMASKLGYFPQKFPLTSFRGSPFQEKMTAGDNPQLVPADQVNITEFYNKSLSSFKENEENIQCGENFMDIECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCRPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKDSRNVFLFKLGGVTASFTASVGSLFLTAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFPHIDETYLMFWIGVTSVLLLFIVYAYMYILWKAHSHAVRMIQRGTQKSIIIHTSEDGKVQVTRPDQARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMFPSCEGTAQPLDNSMGDSDCLHKHANNAASVHRAAESCIKSTVKIAKVTMSVSTDTSAEAL |
| UniProt | P21554 |
| Protein Data Bank | 5tjv, 5u09, 5xr8, 5xra, 6n4b, 5tgz |
| GPCR-HGmod model | P21554 |
| 3D structure model | This structure is from PDB ID 5tjv. |
| BioLiP | BL0384680, BL0364157, BL0384679, BL0384681, BL0384682, BL0384683, BL0384684, BL0440253, BL0440254,BL0440255, BL0363267, BL0361447, BL0361446 |
| Therapeutic Target Database | T76685 |
| ChEMBL | CHEMBL218 |
| IUPHAR | 56 |
| DrugBank | BE0000061 |
| Name | Org 27569 |
|---|---|
| Molecular formula | C24H28ClN3O |
| IUPAC name | 5-chloro-3-ethyl-N-[2-(4-piperidin-1-ylphenyl)ethyl]-1H-indole-2-carboxamide |
| Molecular weight | 409.958 |
| Hydrogen bond acceptor | 2 |
| Hydrogen bond donor | 2 |
| XlogP | 6.0 |
| Synonyms | J-517342 Org 27569, >=98% (HPLC) SW219411-1 5-Chloro-3-ethyl-N-[2-[4-(1-piperidinyl)phenyl]ethyl-1H-indole-2-carboxamide API0008465 [ Show all ] |
| Inchi Key | AHFZDNYNXFMRFQ-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C24H28ClN3O/c1-2-20-21-16-18(25)8-11-22(21)27-23(20)24(29)26-13-12-17-6-9-19(10-7-17)28-14-4-3-5-15-28/h6-11,16,27H,2-5,12-15H2,1H3,(H,26,29) |
| PubChem CID | 44828492 |
| ChEMBL | CHEMBL1553629 |
| IUPHAR | 7851 |
| BindingDB | 50389939 |
| DrugBank | N/A |
Structure | 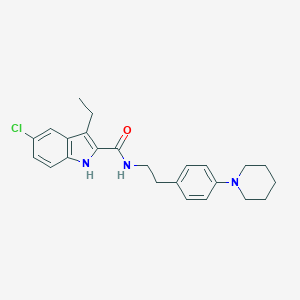 |
| Lipinski's druglikeness | This ligand has a partition coefficient log P greater than 5. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| Activity | >30.0 % | PMID25797163 | ChEMBL |
| Activity | 243.8 % | PMID22571451 | ChEMBL |
| EC50 | 9.05 nM | PMID26529344 | ChEMBL |
| EC50 | 83.0 nM | PMID26529344 | ChEMBL |
| EC50 | 140.0 nM | PMID22571451 | BindingDB,IUPHAR,ChEMBL |
| EC50 | 324.0 nM | PMID26529344 | ChEMBL |
| EC50 | 640.0 nM | PMID26203658 | BindingDB,ChEMBL |
| EC50 | 2090.0 nM | PMID26203658 | BindingDB,ChEMBL |
| EC50 | 2240.0 nM | PMID26203658 | BindingDB,ChEMBL |
| EC50 | 10590.0 nM | PMID26203658 | BindingDB,ChEMBL |
| Emax | 78.1 % | PMID26529344 | ChEMBL |
| Emax | 105.0 % | PMID26529344 | ChEMBL |
| Emax | 109.3 % | PMID26529344 | ChEMBL |
| IC50 | 853.0 nM | PMID25797163 | BindingDB,ChEMBL |
| Kb | 217.3 nM | PMID24053617, PMID24635495 | ChEMBL |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218