

You can:
| Name | Metabotropic glutamate receptor 8 |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Grm8 |
| Synonym | GLUR8 glutamate receptor Gprc1h mGlu8 receptor mGluR8 [ Show all ] |
| Disease | N/A for non-human GPCRs |
| Length | 908 |
| Amino acid sequence | MVCEGKRLASCPCFFLLTAKFYWILTMMQRTHSQEYAHSIRVDGDIILGGLFPVHAKGERGVPCGELKKEKGIHRLEAMLYAIDQINKDPDLLSNITLGVRILDTCSRDTYALEQSLTFVQALIEKDASDVKCANGDPPIFTKPDKISGVIGAAASSVSIMVANILRLFKIPQISYASTAPELSDNTRYDFFSRVVPPDSYQAQAMVDIVTALGWNYVSTLASEGNYGESGVEAFTQISREIGGVCIAQSQKIPREPRPGEFEKIIKRLLETPNARAVIMFANEDDIRRILEAAKKLNQSGHFLWIGSDSWGSKIAPVYQQEEIAEGAVTILPKRASIDGFDRYFRSRTLANNRRNVWFAEFWEENFGCKLGSHGKRNSHIKKCTGLERIARDSSYEQEGKVQFVIDAVYSMAYALHNMHKERCPGYIGLCPRMVTIDGKELLGYIRAVNFNGSAGTPVTFNENGDAPGRYDIFQYQINNKSTEYKIIGHWTNQLHLKVEDMQWANREHTHPASVCSLPCKPGERKKTVKGVPCCWHCERCEGYNYQVDELSCELCPLDQRPNINRTGCQRIPIIKLEWHSPWAVVPVFIAILGIIATTFVIVTFVRYNDTPIVRASGRELSYVLLTGIFLCYSITFLMIAAPDTIICSFRRIFLGLGMCFSYAALLTKTNRIHRIFEQGKKSVTAPKFISPASQLVITFSLISVQLLGVFVWFVVDPPHTIIDYGEQRTLDPENARGVLKCDISDLSLICSLGYSILLMVTCTVYAIKTRGVPETFNEAKPIGFTMYTTCIIWLAFIPIFFGTAQSAEKMYIQTTTLTVSMSLSASVSLGMLYMPKVYIIIFHPEQNVQKRKRSFKAVVTAATMQSKLIQKGNDRPNGEVKSELCESLETNTSSTKTTYISYSNHSI |
| UniProt | P70579 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL2718 |
| IUPHAR | 296 |
| DrugBank | N/A |
| Name | L-AP4 |
|---|---|
| Molecular formula | C4H10NO5P |
| IUPAC name | (2S)-2-amino-4-phosphonobutanoic acid |
| Molecular weight | 183.1 |
| Hydrogen bond acceptor | 6 |
| Hydrogen bond donor | 4 |
| XlogP | -5.5 |
| Synonyms | (2S)-2-amino-4-phosphonobutanoic acid 23052-81-5 BN0084 D0H7UP J-014961 [ Show all ] |
| Inchi Key | DDOQBQRIEWHWBT-VKHMYHEASA-N |
| Inchi ID | InChI=1S/C4H10NO5P/c5-3(4(6)7)1-2-11(8,9)10/h3H,1-2,5H2,(H,6,7)(H2,8,9,10)/t3-/m0/s1 |
| PubChem CID | 179394 |
| ChEMBL | CHEMBL33567 |
| IUPHAR | 1410, 1412 |
| BindingDB | 50007548 |
| DrugBank | N/A |
Structure | 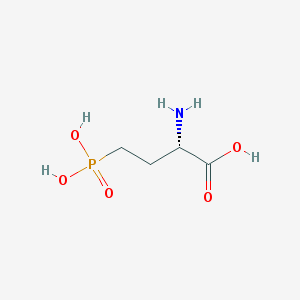 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| EC50 | 60.0 nM | PMID14711315 | BindingDB,ChEMBL |
| EC50 | 70.0 nM | PMID26814576 | BindingDB,ChEMBL |
| EC50 | 110.0 nM | PMID25958247 | BindingDB,ChEMBL |
| EC50 | 128.0 nM | PMID17722902 | BindingDB,ChEMBL |
| EC50 | 240.0 nM | PMID20218620 | BindingDB,ChEMBL |
| EC50 | 300.0 nM | PMID20218620 | BindingDB,ChEMBL |
| EC50 | 630.957 nM | PMID9016353 | IUPHAR |
| Ki | 1900.0 nM | PMID17350267, PMID16213710 | BindingDB,ChEMBL |
| Ki | 2700.0 nM | PMID11114395 | BindingDB |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218