

You can:
| Name | Mu-type opioid receptor |
|---|---|
| Species | Mus musculus (Mouse) |
| Gene | Oprm1 |
| Synonym | opioid receptor, mu 1 opioid receptor OP3 Mu receptor MOR-1 [ Show all ] |
| Disease | N/A for non-human GPCRs |
| Length | 398 |
| Amino acid sequence | MDSSAGPGNISDCSDPLAPASCSPAPGSWLNLSHVDGNQSDPCGPNRTGLGGSHSLCPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALATSTLPFQSVNYLMGTWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYRQGSIDCTLTFSHPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGSKEKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALITIPETTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCFREFCIPTSSTIEQQNSARIRQNTREHPSTANTVDRTNHQLENLEAETAPLP |
| UniProt | P42866 |
| Protein Data Bank | 4dkl, 5c1m, 6dde, 6ddf |
| GPCR-HGmod model | N/A |
| 3D structure model | This structure is from PDB ID 4dkl. |
| BioLiP | BL0416752, BL0416751, BL0321492, BL0224753, BL0224754, BL0224755,BL0224756, BL0321491 |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL2858 |
| IUPHAR | 319 |
| DrugBank | N/A |
| Name | naltrexone |
|---|---|
| Molecular formula | C20H23NO4 |
| IUPAC name | (4R,4aS,7aR,12bS)-3-(cyclopropylmethyl)-4a,9-dihydroxy-2,4,5,6,7a,13-hexahydro-1H-4,12-methanobenzofuro[3,2-e]isoquinolin-7-one |
| Molecular weight | 341.407 |
| Hydrogen bond acceptor | 5 |
| Hydrogen bond donor | 2 |
| XlogP | 1.9 |
| Synonyms | (4R,4aS,7aR,12bS)-3-(cyclopropylmethyl)-4a,9-dihydroxy-2,4,5,6,7a,13-hexahydro-1H-4,12-methanobenzofuro[3,2-e]isoquinoline-7-one Naltrexone (USAN/INN) Naltrexone [USAN:INN:BAN] AB00174152-14 Opioid antagonists (irritable bowel syndrome), Pain Therapeutics [ Show all ] |
| Inchi Key | DQCKKXVULJGBQN-XFWGSAIBSA-N |
| Inchi ID | InChI=1S/C20H23NO4/c22-13-4-3-12-9-15-20(24)6-5-14(23)18-19(20,16(12)17(13)25-18)7-8-21(15)10-11-1-2-11/h3-4,11,15,18,22,24H,1-2,5-10H2/t15-,18+,19+,20-/m1/s1 |
| PubChem CID | 5360515 |
| ChEMBL | CHEMBL19019 |
| IUPHAR | 1639 |
| BindingDB | 50000787, 60212 |
| DrugBank | DB00704 |
Structure | 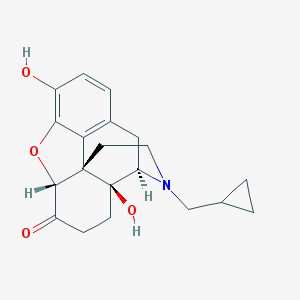 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| AD50 | 0.05 mg kg-1 | PMID8410998, PMID8410999 | ChEMBL |
| Dose ratio | 50.2 - | PMID2409281 | ChEMBL |
| EC50 | 0.16 nM | PMID23948248, PMID25783191 | ChEMBL |
| EC50 | 0.16 nM | PMID25783191 | BindingDB |
| Emax | 5.4 % | PMID23948248, PMID25783191 | ChEMBL |
| Kd | 1.738 nM | PMID17962026 | ChEMBL |
| Ke | 0.09 nM | PMID18062664 | ChEMBL |
| Ke | 0.21 nM | PMID14736254 | ChEMBL |
| Ke | 0.24 nM | PMID18062664 | ChEMBL |
| Ke | 0.44 nM | PMID19253983 | ChEMBL |
| Ke | 0.57 nM | PMID18062664 | ChEMBL |
| Ki | 0.265 nM | PMID21889335 | ChEMBL |
| Ki | 0.33 nM | PMID25783191 | BindingDB |
| Ki | 0.33 nM | PMID23948248, PMID25783191 | ChEMBL |
| Ki | 0.6457 nM | PMID23585918 | ChEMBL |
| Ki | 0.646 nM | PMID23585918 | BindingDB |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218