

You can:
| Name | Galanin receptor type 3 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | GALR3 |
| Synonym | GAL3-R Galnr3 GALR-3 GALR3 GAL3 receptor |
| Disease | Cognitive disorders |
| Length | 368 |
| Amino acid sequence | MADAQNISLDSPGSVGAVAVPVVFALIFLLGTVGNGLVLAVLLQPGPSAWQEPGSTTDLFILNLAVADLCFILCCVPFQATIYTLDAWLFGALVCKAVHLLIYLTMYASSFTLAAVSVDRYLAVRHPLRSRALRTPRNARAAVGLVWLLAALFSAPYLSYYGTVRYGALELCVPAWEDARRRALDVATFAAGYLLPVAVVSLAYGRTLRFLWAAVGPAGAAAAEARRRATGRAGRAMLAVAALYALCWGPHHALILCFWYGRFAFSPATYACRLASHCLAYANSCLNPLVYALASRHFRARFRRLWPCGRRRRHRARRALRRVRPASSGPPGCPGDARPSGRLLAGGGQGPEPREGPVHGGEAARGPE |
| UniProt | O60755 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | O60755 |
| 3D structure model | This predicted structure model is from GPCR-EXP O60755. |
| BioLiP | N/A |
| Therapeutic Target Database | T98494 |
| ChEMBL | CHEMBL2731 |
| IUPHAR | 245 |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 106 | 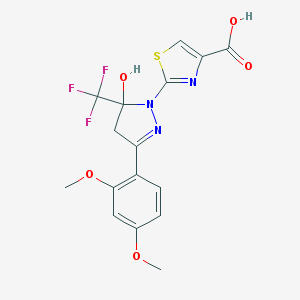 MLS000716936 MLS000716936 | C16H14F3N3O5S | 417.359 | 12 / 2 | 3.1 | No |
| 415 |  Dibromosalicil Dibromosalicil | C14H8Br2O4 | 400.022 | 4 / 2 | 4.7 | Yes |
| 5352 | 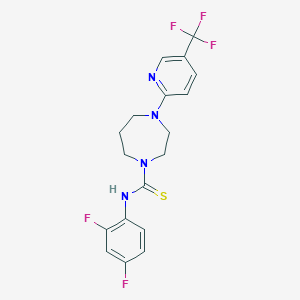 MLS000833548 MLS000833548 | C18H17F5N4S | 416.414 | 8 / 1 | 4.0 | Yes |
| 5954 | 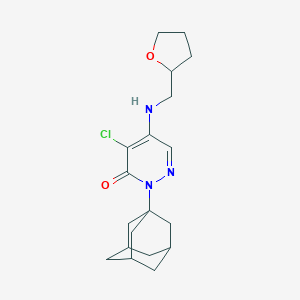 MLS000730667 MLS000730667 | C19H26ClN3O2 | 363.886 | 4 / 1 | 3.6 | Yes |
| 6260 | 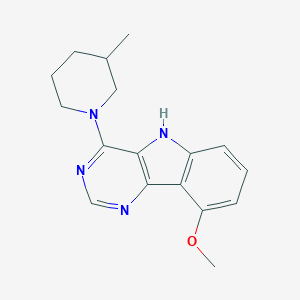 MLS000040609 MLS000040609 | C17H20N4O | 296.374 | 4 / 1 | 3.5 | Yes |
| 7062 | 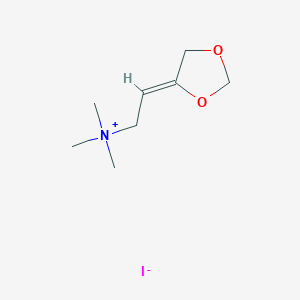 SMR000033717 SMR000033717 | C8H16INO2 | 285.125 | 3 / 0 | N/A | N/A |
| 7363 | 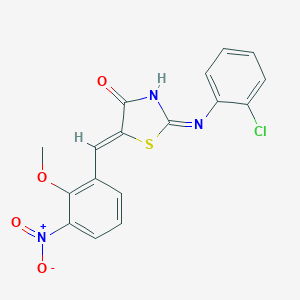 MLS001209947 MLS001209947 | C17H12ClN3O4S | 389.81 | 6 / 1 | 4.5 | Yes |
| 8822 | 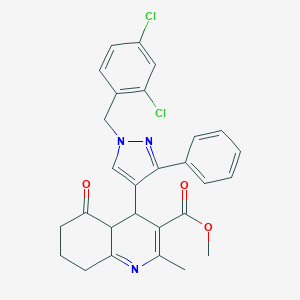 BDBM97075 BDBM97075 | C28H25Cl2N3O3 | 522.426 | 5 / 0 | 4.8 | No |
| 9987 | 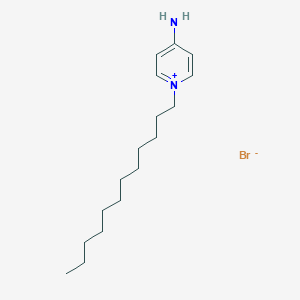 MolPort-000-723-526 MolPort-000-723-526 | C17H31BrN2 | 343.353 | 2 / 1 | N/A | N/A |
| 14458 | 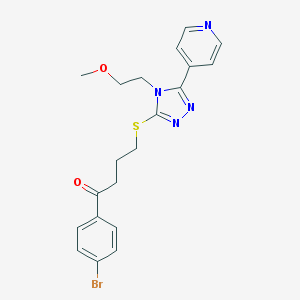 MLS000566658 MLS000566658 | C20H21BrN4O2S | 461.378 | 6 / 0 | 3.0 | Yes |
| 14500 | 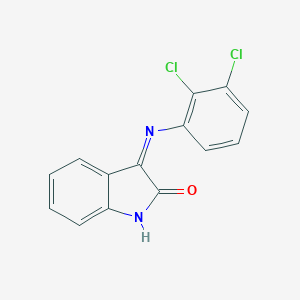 CHEMBL378877 CHEMBL378877 | C14H8Cl2N2O | 291.131 | 2 / 1 | 3.8 | Yes |
| 21492 | 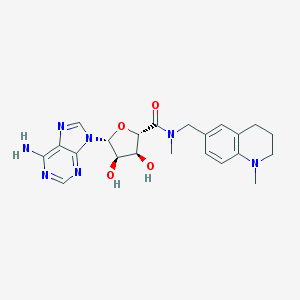 MLS003271268 MLS003271268 | C22H27N7O4 | 453.503 | 9 / 3 | 0.3 | Yes |
| 22951 | 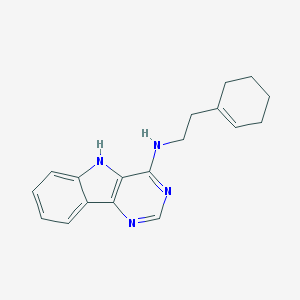 SMR000220470 SMR000220470 | C18H20N4 | 292.386 | 3 / 2 | 4.1 | Yes |
| 28407 | 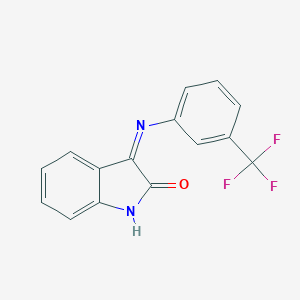 CHEMBL209480 CHEMBL209480 | C15H9F3N2O | 290.245 | 5 / 1 | 3.4 | Yes |
| 29037 |  MLS000538535 MLS000538535 | C15H15BrN2O2 | 335.201 | 3 / 1 | 3.1 | Yes |
| 30772 | 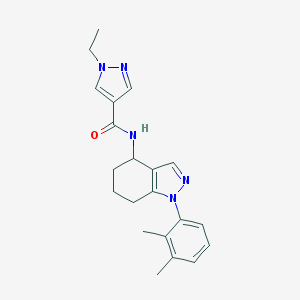 MLS001125299 MLS001125299 | C21H25N5O | 363.465 | 3 / 1 | 2.8 | Yes |
| 31592 | 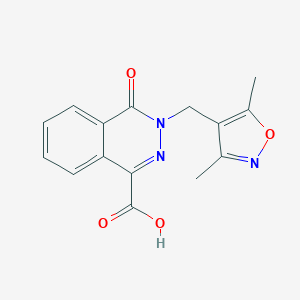 MLS000054201 MLS000054201 | C15H13N3O4 | 299.286 | 6 / 1 | 1.6 | Yes |
| 33982 | 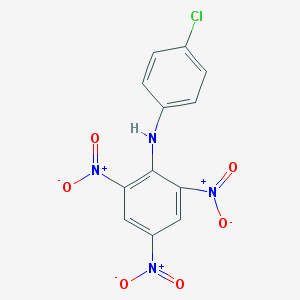 MLS002920666 MLS002920666 | C12H7ClN4O6 | 338.66 | 7 / 1 | 4.1 | Yes |
| 35827 | 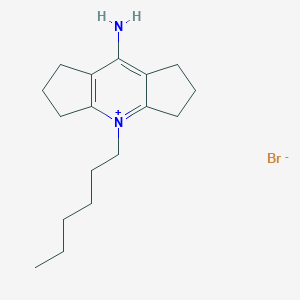 MolPort-000-720-960 MolPort-000-720-960 | C17H27BrN2 | 339.321 | 2 / 1 | N/A | N/A |
| 38174 | 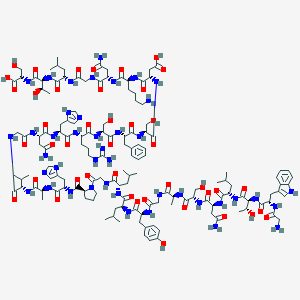 CHEMBL506553 CHEMBL506553 | C139H210N42O43 | 3157.46 | 48 / 48 | -15.9 | No |
| 39853 | 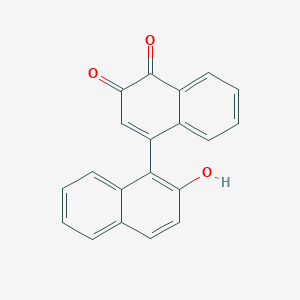 MLS002701634 MLS002701634 | C20H12O3 | 300.313 | 3 / 1 | 3.6 | Yes |
| 41874 |  MLS000040640 MLS000040640 | C18H25N5O2 | 343.431 | 6 / 1 | 4.2 | Yes |
| 42613 |  MLS000519909 MLS000519909 | C26H30N4O5 | 478.549 | 6 / 1 | 3.0 | Yes |
| 43898 | 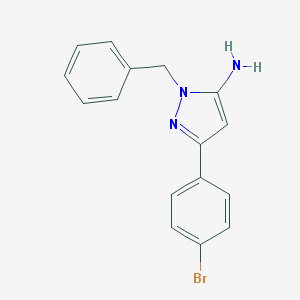 MLS000712366 MLS000712366 | C16H14BrN3 | 328.213 | 2 / 1 | 3.8 | Yes |
| 44948 |  SMR000001597 SMR000001597 | C19H16N4O3S | 380.422 | 7 / 0 | 3.1 | Yes |
| 46945 |  MLS002253020 MLS002253020 | C20H18N2O3 | 334.375 | 4 / 1 | 3.6 | Yes |
| 47560 |  CHEMBL209737 CHEMBL209737 | C14H9ClN2O | 256.689 | 2 / 1 | 3.3 | Yes |
| 48892 |  MLS001000456 MLS001000456 | C17H19N3O2S | 329.418 | 6 / 1 | 4.2 | Yes |
| 51013 |  MLS002636314 MLS002636314 | C23H17F2N3O4S2 | 501.523 | 9 / 2 | 5.0 | No |
| 52651 |  MLS001002564 MLS001002564 | C17H14N2O4 | 310.309 | 5 / 1 | 2.3 | Yes |
| 54795 |  MLS001177315 MLS001177315 | C25H26N2O4 | 418.493 | 4 / 1 | 3.8 | Yes |
| 55934 |  MLS002164468 MLS002164468 | C20H28N6OS | 400.545 | 6 / 0 | 2.8 | Yes |
| 56610 | 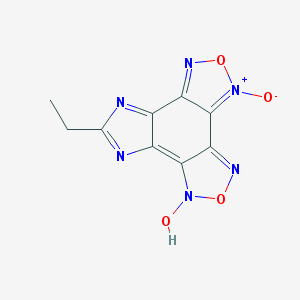 MLS000094981 MLS000094981 | C9H6N6O4 | 262.185 | 8 / 1 | 0.9 | Yes |
| 58524 | 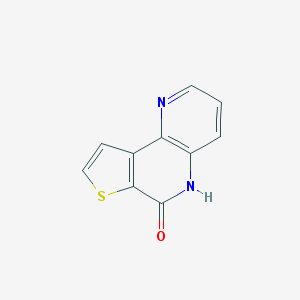 MLS000704511 MLS000704511 | C10H6N2OS | 202.231 | 3 / 1 | 1.4 | Yes |
| 559107 | 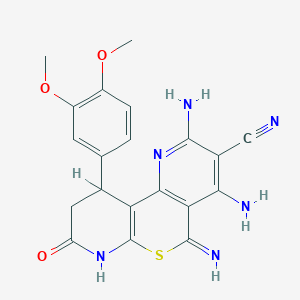 SMR000147834 SMR000147834 | C20H18N6O3S | 422.463 | 9 / 4 | 1.0 | Yes |
| 60392 | 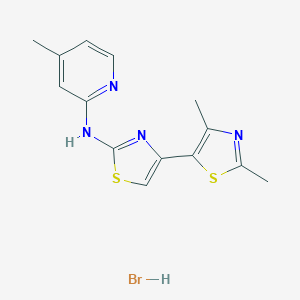 MLS001180182 MLS001180182 | C14H15BrN4S2 | 383.326 | 6 / 2 | N/A | N/A |
| 60847 |  SMR000010338 SMR000010338 | C15H14ClN3 | 271.748 | 3 / 2 | N/A | N/A |
| 559180 |  MLS000766664 MLS000766664 | C14H11BrN4O5 | 395.169 | 7 / 2 | 1.9 | Yes |
| 559187 |  SMR000147833 SMR000147833 | C18H14N6OS | 362.411 | 7 / 4 | 1.1 | Yes |
| 63444 |  MLS001029145 MLS001029145 | C29H38ClN7O | 536.121 | 6 / 1 | 4.3 | No |
| 63830 |  MLS000082037 MLS000082037 | C24H23N3O4 | 417.465 | 5 / 0 | 4.0 | Yes |
| 64028 |  MLS001030649 MLS001030649 | C25H28ClN3O2S | 470.028 | 5 / 1 | 4.9 | Yes |
| 64847 |  NSC720623 NSC720623 | C25H26F3N3O3S | 505.556 | 7 / 0 | N/A | No |
| 67485 |  MLS001174949 MLS001174949 | C18H22N2O3 | 314.385 | 4 / 1 | 3.4 | Yes |
| 68273 | 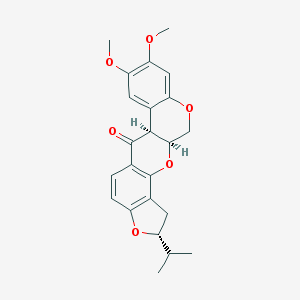 Dihydrorotenone Dihydrorotenone | C23H24O6 | 396.439 | 6 / 0 | 4.2 | Yes |
| 68274 |  Dihydrorotenone Dihydrorotenone | C23H24O6 | 396.439 | 6 / 0 | 4.2 | Yes |
| 70542 | 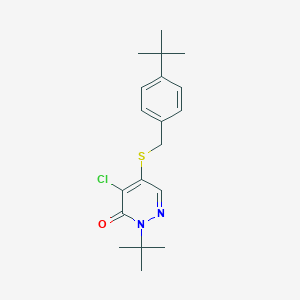 Pyridaben Pyridaben | C19H25ClN2OS | 364.932 | 3 / 0 | 5.2 | No |
| 73261 | 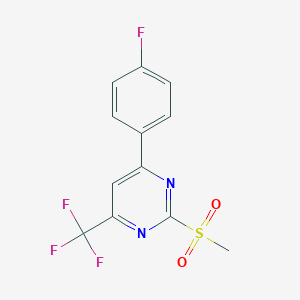 MLS000675919 MLS000675919 | C12H8F4N2O2S | 320.262 | 8 / 0 | 2.5 | Yes |
| 74560 | 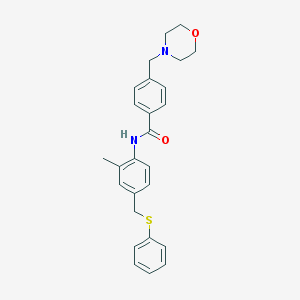 MLS000664008 MLS000664008 | C26H28N2O2S | 432.582 | 4 / 1 | 4.6 | Yes |
| 74702 |  MLS000571626 MLS000571626 | C13H16N4O3 | 276.296 | 5 / 1 | 1.4 | Yes |
| 74905 |  2,6-Di-tert-butyl-4-nitrophenol 2,6-Di-tert-butyl-4-nitrophenol | C14H21NO3 | 251.326 | 3 / 1 | 4.7 | Yes |
| 76533 |  SMR000006308 SMR000006308 | C17H26N4O4S | 382.479 | 7 / 3 | 2.8 | Yes |
| 78688 |  Galanin-1-16 Galanin-1-16 | C78H116N20O21 | 1669.91 | 23 / 22 | -2.9 | No |
| 79213 |  AC1M530P AC1M530P | C23H23N2OS+ | 375.51 | 3 / 0 | 5.9 | No |
| 81875 |  SMR000122163 SMR000122163 | C21H35BrN2 | 395.429 | 2 / 2 | N/A | N/A |
| 82740 | 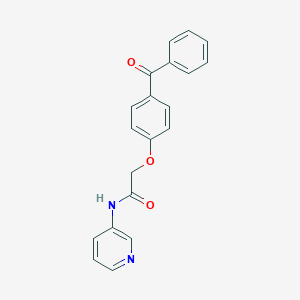 MLS001179913 MLS001179913 | C20H16N2O3 | 332.359 | 4 / 1 | 3.1 | Yes |
| 87709 |  MLS000732847 MLS000732847 | C23H25N3O2 | 375.472 | 4 / 1 | 3.9 | Yes |
| 88578 | 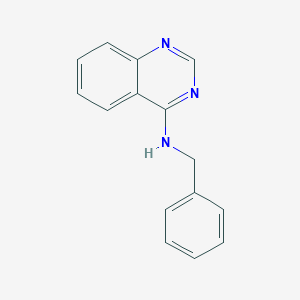 N-Benzylquinazolin-4-amine N-Benzylquinazolin-4-amine | C15H13N3 | 235.29 | 3 / 1 | 3.9 | Yes |
| 89210 | 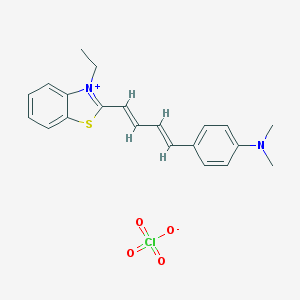 Exciton Exciton | C21H23ClN2O4S | 434.935 | 6 / 0 | N/A | N/A |
| 91049 | 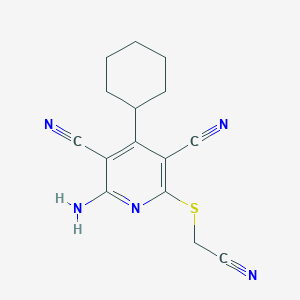 MLS000085820 MLS000085820 | C15H15N5S | 297.38 | 6 / 1 | 3.7 | Yes |
| 93188 | 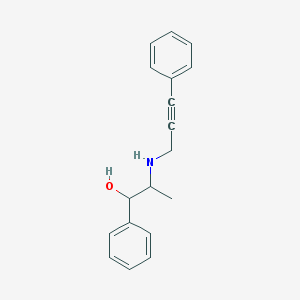 MLS000684433 MLS000684433 | C18H19NO | 265.356 | 2 / 2 | 3.0 | Yes |
| 96859 | 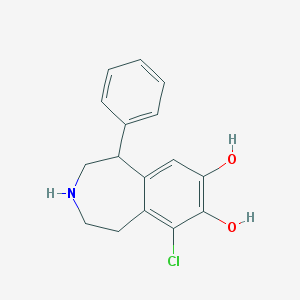 Skf-81297 Skf-81297 | C16H16ClNO2 | 289.759 | 3 / 3 | 2.7 | Yes |
| 96954 | 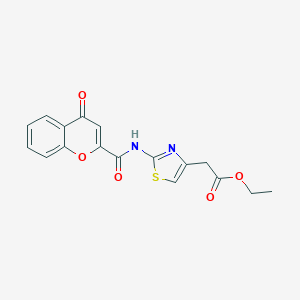 MLS000683121 MLS000683121 | C17H14N2O5S | 358.368 | 7 / 1 | 2.1 | Yes |
| 96994 | 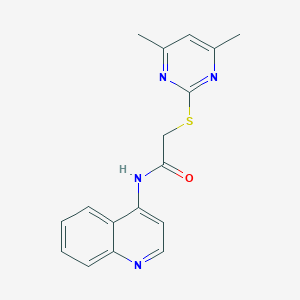 MLS000662478 MLS000662478 | C17H16N4OS | 324.402 | 5 / 1 | 3.0 | Yes |
| 97214 | 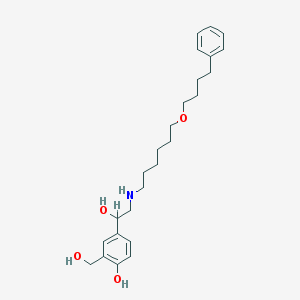 Salmeterol Salmeterol | C25H37NO4 | 415.574 | 5 / 4 | 3.9 | Yes |
| 97382 | 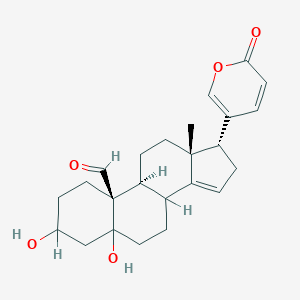 MLS001215955 MLS001215955 | C24H30O5 | 398.499 | 5 / 2 | 2.4 | Yes |
| 99173 |  MLS003119183 MLS003119183 | C19H18N2O4S | 370.423 | 6 / 2 | 2.7 | Yes |
| 99910 | 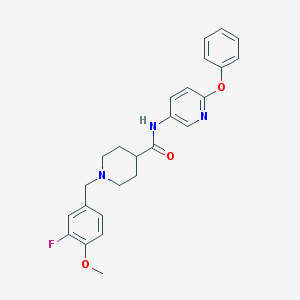 MLS001100829 MLS001100829 | C25H26FN3O3 | 435.499 | 6 / 1 | 4.0 | Yes |
| 100845 | 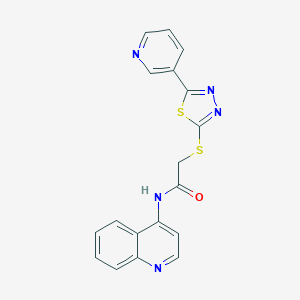 SMR000221403 SMR000221403 | C18H13N5OS2 | 379.456 | 7 / 1 | 3.0 | Yes |
| 101585 |  MLS000998903 MLS000998903 | C21H30FN3OS | 391.549 | 6 / 1 | 3.9 | Yes |
| 101827 |  2-ethyl-N-(5-nitropyridin-2-yl)butanamide 2-ethyl-N-(5-nitropyridin-2-yl)butanamide | C11H15N3O3 | 237.259 | 4 / 1 | 1.9 | Yes |
| 104108 |  BDBM70384 BDBM70384 | C23H21N5O2 | 399.454 | 5 / 2 | 3.2 | Yes |
| 104132 |  MLS001180182 MLS001180182 | C14H14N4S2 | 302.414 | 6 / 1 | 4.0 | Yes |
| 106315 |  CHEMBL1323872 CHEMBL1323872 | C23H23IN2OS | 502.414 | 4 / 0 | N/A | No |
| 106462 |  CHEMBL1588926 CHEMBL1588926 | C25H25BrN2 | 433.393 | 2 / 0 | N/A | N/A |
| 107720 |  MLS-0380219.0001 MLS-0380219.0001 | C18H17F2N5O | 357.365 | 5 / 1 | 2.3 | Yes |
| 560937 |  SMR000060129 SMR000060129 | C19H20N2O5 | 356.378 | 6 / 3 | 3.8 | Yes |
| 118389 | 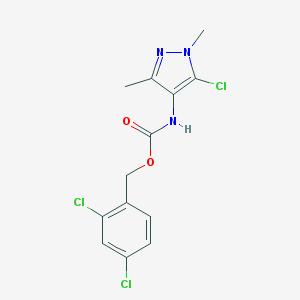 MLS000547376 MLS000547376 | C13H12Cl3N3O2 | 348.608 | 3 / 1 | 3.9 | Yes |
| 123281 |  MLS002249163 MLS002249163 | C15H16N4O5S2 | 396.436 | 8 / 1 | 1.4 | Yes |
| 123504 | 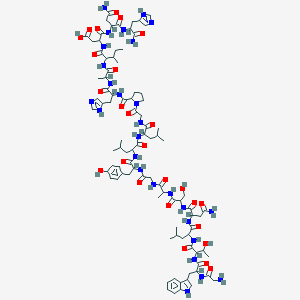 Galanin rat Galanin rat | C92H135N27O26 | 2035.26 | 29 / 28 | -6.9 | No |
| 125723 | 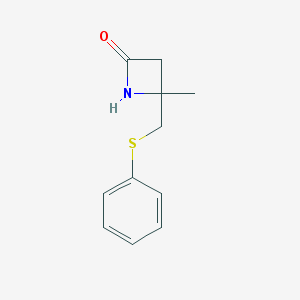 MLS003115736 MLS003115736 | C11H13NOS | 207.291 | 2 / 1 | 1.7 | Yes |
| 553920 | 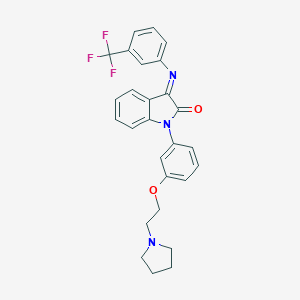 SNAP-398299 SNAP-398299 | C27H24F3N3O2 | 479.503 | 7 / 0 | 5.7 | No |
| 127210 |  MLS000701132 MLS000701132 | C13H13Cl2N3O | 298.167 | 2 / 1 | 3.0 | Yes |
| 127993 |  MLS001007488 MLS001007488 | C16H17N3O2S | 315.391 | 6 / 1 | 3.3 | Yes |
| 130599 |  AKOS015896602 AKOS015896602 | C17H22ClNO3 | 323.817 | 5 / 5 | N/A | N/A |
| 131806 |  BAS 00918756 BAS 00918756 | C7H11ClN4O2S | 250.701 | 6 / 3 | 0.8 | Yes |
| 134279 |  MLS000391427 MLS000391427 | C21H18Cl2N2O3S | 449.346 | 5 / 1 | 5.4 | No |
| 138246 |  ritodrine ritodrine | C17H21NO3 | 287.359 | 4 / 4 | 2.3 | Yes |
| 138991 |  MLS001160878 MLS001160878 | C20H23N3O5 | 385.42 | 6 / 1 | 2.7 | Yes |
| 138992 |  AKOS033707998 AKOS033707998 | C20H23N3O5 | 385.42 | 6 / 1 | 2.7 | Yes |
| 139967 |  MLS000717757 MLS000717757 | C19H30N2 | 286.463 | 2 / 1 | 5.1 | No |
| 141336 | 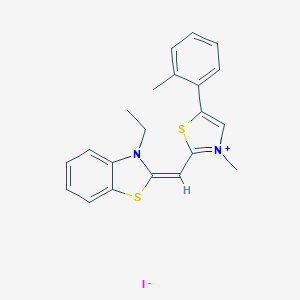 CID 73950903 CID 73950903 | C21H21IN2S2 | 492.437 | 4 / 0 | N/A | N/A |
| 144261 |  Bio3H4 Bio3H4 | C13H7F3N6 | 304.236 | 8 / 1 | 3.6 | Yes |
| 144651 | 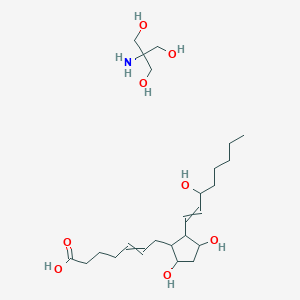 MLS003107056 MLS003107056 | C24H45NO8 | 475.623 | 9 / 8 | N/A | No |
| 145355 | 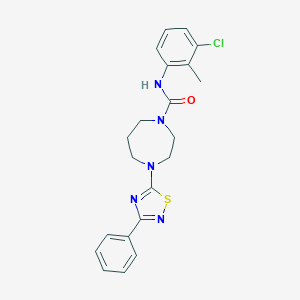 MLS000833557 MLS000833557 | C21H22ClN5OS | 427.951 | 5 / 1 | 4.9 | Yes |
| 148164 | 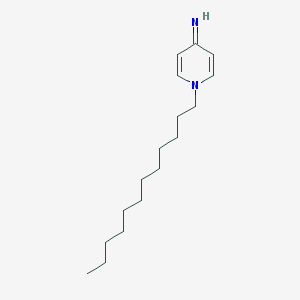 GNF-Pf-2344 GNF-Pf-2344 | C17H30N2 | 262.441 | 2 / 1 | 6.0 | No |
| 149138 |  MLS003231005 MLS003231005 | C19H14BrClN2O | 401.688 | 2 / 0 | 6.2 | No |
| 149359 |  MLS000077260 MLS000077260 | C16H16N4OS2 | 344.451 | 6 / 1 | 3.9 | Yes |
| 149652 |  MLS003120629 MLS003120629 | C23H20N2O4 | 388.423 | 5 / 1 | 3.6 | Yes |
| 150640 |  MLS003230991 MLS003230991 | C23H15F3N2O | 392.381 | 5 / 0 | 6.6 | No |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218