

You can:
| Name | C5a anaphylatoxin chemotactic receptor 1 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | C5AR1 |
| Synonym | complement component 5a receptor 1 complement C5a receptor C5A CD88 C5R1 [ Show all ] |
| Disease | Vasculitis Atopic dermatitis Inflammatory disease Rheumatoid arthritis |
| Length | 350 |
| Amino acid sequence | MDSFNYTTPDYGHYDDKDTLDLNTPVDKTSNTLRVPDILALVIFAVVFLVGVLGNALVVWVTAFEAKRTINAIWFLNLAVADFLSCLALPILFTSIVQHHHWPFGGAACSILPSLILLNMYASILLLATISADRFLLVFKPIWCQNFRGAGLAWIACAVAWGLALLLTIPSFLYRVVREEYFPPKVLCGVDYSHDKRRERAVAIVRLVLGFLWPLLTLTICYTFILLRTWSRRATRSTKTLKVVVAVVASFFIFWLPYQVTGIMMSFLEPSSPTFLLLKKLDSLCVSFAYINCCINPIIYVVAGQGFQGRLRKSLPSLLRNVLTEESVVRESKSFTRSTVDTMAQKTQAV |
| UniProt | P21730 |
| Protein Data Bank | 6c1r, 6c1q, 5o9h |
| GPCR-HGmod model | P21730 |
| 3D structure model | This structure is from PDB ID 6c1r. |
| BioLiP | BL0415511, BL0415514, BL0415513, BL0415512, BL0401194,BL0401195,BL0401196, |
| Therapeutic Target Database | T15439 |
| ChEMBL | CHEMBL2373 |
| IUPHAR | 32 |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 2369 | 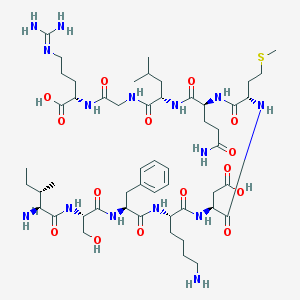 CHEMBL441393 CHEMBL441393 | C52H87N15O15S | 1194.42 | 19 / 17 | -7.6 | No |
| 2417 | 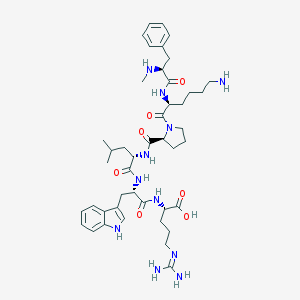 CHEMBL117751 CHEMBL117751 | C44H65N11O7 | 860.074 | 10 / 10 | -0.6 | No |
| 3823 | 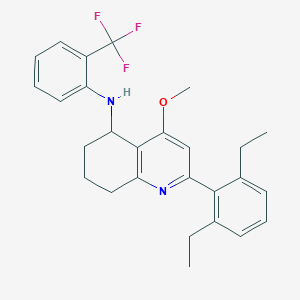 CHEMBL481755 CHEMBL481755 | C27H29F3N2O | 454.537 | 6 / 1 | 7.3 | No |
| 4424 | 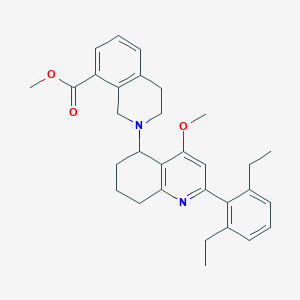 CHEMBL261594 CHEMBL261594 | C31H36N2O3 | 484.64 | 5 / 0 | 6.1 | No |
| 4925 |  CHEMBL435777 CHEMBL435777 | C45H63N11O6 | 854.07 | 8 / 9 | 3.0 | No |
| 4926 | 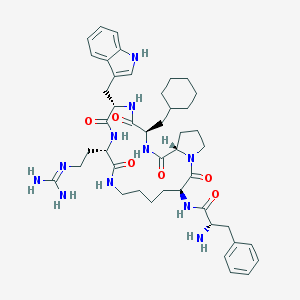 CHEMBL293241 CHEMBL293241 | C45H63N11O6 | 854.07 | 8 / 9 | 3.0 | No |
| 5877 | 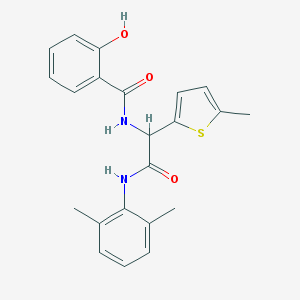 diamide 7 diamide 7 | C22H22N2O3S | 394.489 | 4 / 3 | 4.8 | Yes |
| 5921 | 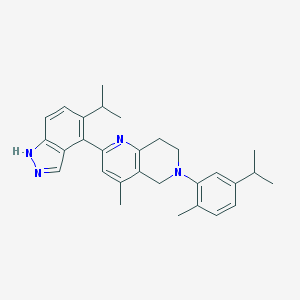 CHEMBL3646994 CHEMBL3646994 | C29H34N4 | 438.619 | 3 / 1 | 6.8 | No |
| 6365 | 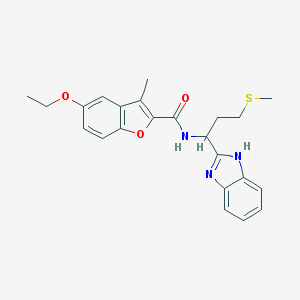 methionine benzimidazole 6 methionine benzimidazole 6 | C23H25N3O3S | 423.531 | 5 / 2 | 4.8 | Yes |
| 8632 | 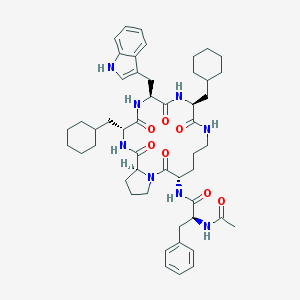 CHEMBL425289 CHEMBL425289 | C50H68N8O7 | 893.143 | 7 / 7 | 7.2 | No |
| 8821 | 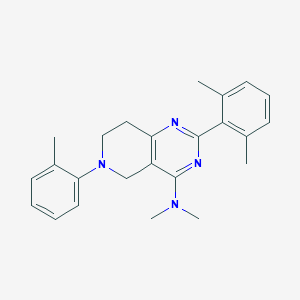 CHEMBL3646909 CHEMBL3646909 | C24H28N4 | 372.516 | 4 / 0 | 5.1 | No |
| 9047 | 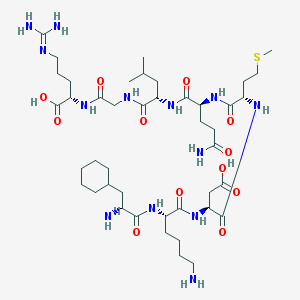 CHEMBL339127 CHEMBL339127 | C43H77N13O12S | 1000.23 | 16 / 14 | -6.2 | No |
| 459300 | 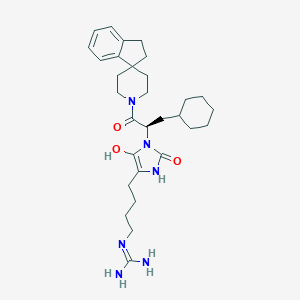 BDBM50290481 BDBM50290481 | C30H44N6O3 | 536.721 | 4 / 4 | 4.1 | No |
| 10634 | 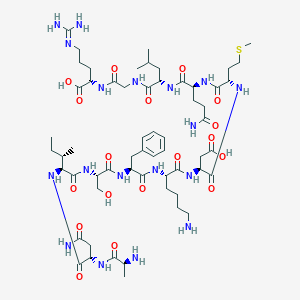 CHEMBL408929 CHEMBL408929 | C59H98N18O18S | 1379.6 | 22 / 20 | -9.6 | No |
| 10821 | 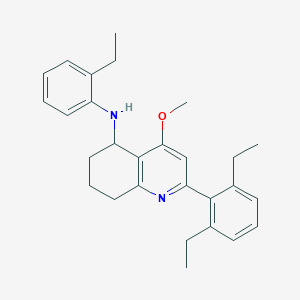 CHEMBL479540 CHEMBL479540 | C28H34N2O | 414.593 | 3 / 1 | 7.2 | No |
| 10841 | 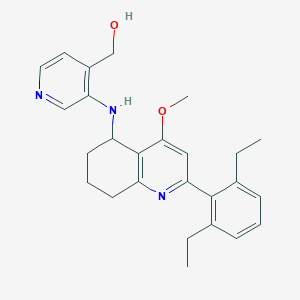 CHEMBL479557 CHEMBL479557 | C26H31N3O2 | 417.553 | 5 / 2 | 5.0 | Yes |
| 11281 | 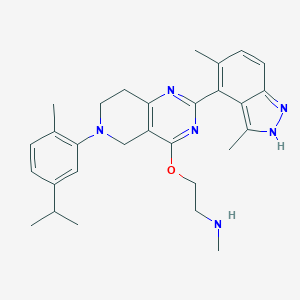 CHEMBL3647016 CHEMBL3647016 | C29H36N6O | 484.648 | 6 / 2 | 5.3 | No |
| 13470 | 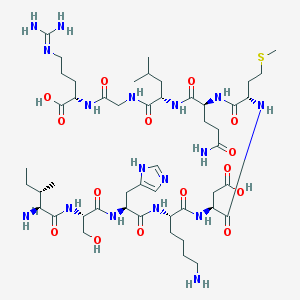 ISHKDMQLGR ISHKDMQLGR | C49H85N17O15S | 1184.38 | 20 / 18 | -9.4 | No |
| 14819 | 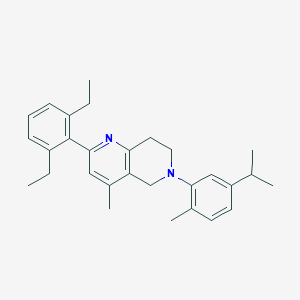 CHEMBL3646995 CHEMBL3646995 | C29H36N2 | 412.621 | 2 / 0 | 7.7 | No |
| 548052 | 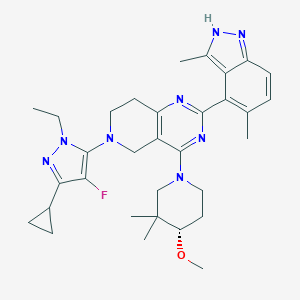 CHEMBL3985025 CHEMBL3985025 | C32H41FN8O | 572.733 | 8 / 1 | 5.3 | No |
| 557682 | 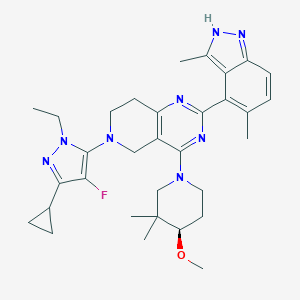 SCHEMBL14663067 SCHEMBL14663067 | C32H41FN8O | 572.733 | 8 / 1 | 5.3 | No |
| 15002 | 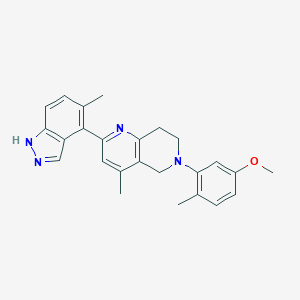 CHEMBL3647034 CHEMBL3647034 | C25H26N4O | 398.51 | 4 / 1 | 4.9 | Yes |
| 15677 | 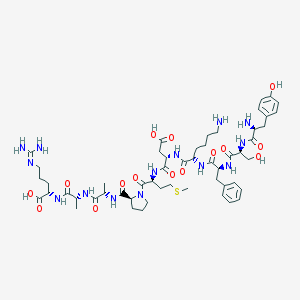 CHEMBL412249 CHEMBL412249 | C53H80N14O15S | 1185.37 | 19 / 16 | -7.1 | No |
| 17343 | 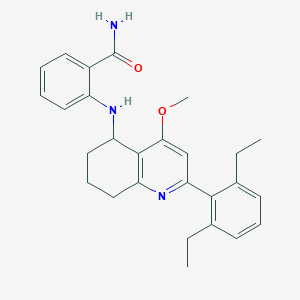 CHEMBL521090 CHEMBL521090 | C27H31N3O2 | 429.564 | 4 / 2 | 5.8 | No |
| 548095 | 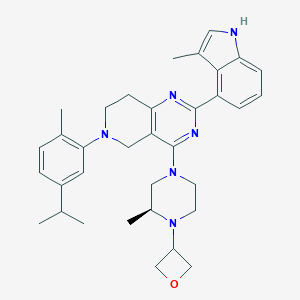 CHEMBL3963795 CHEMBL3963795 | C34H42N6O | 550.751 | 6 / 1 | 5.9 | No |
| 557785 | 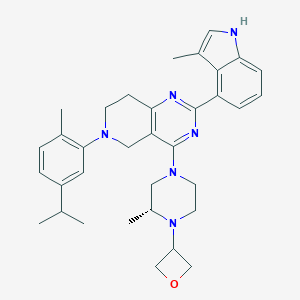 SCHEMBL14663100 SCHEMBL14663100 | C34H42N6O | 550.751 | 6 / 1 | 5.9 | No |
| 18356 | 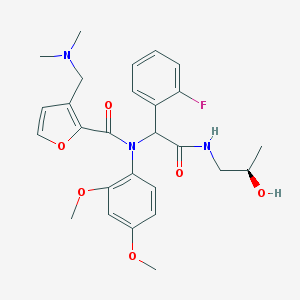 CHEMBL512543 CHEMBL512543 | C27H32FN3O6 | 513.566 | 8 / 2 | 3.0 | No |
| 18477 |  CHEMBL261966 CHEMBL261966 | C58H89N15O16S | 1284.5 | 20 / 17 | -7.0 | No |
| 557828 | 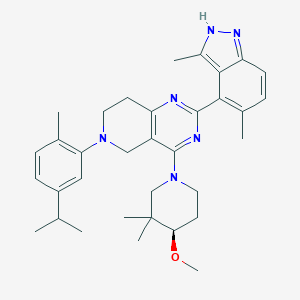 SCHEMBL14671793 SCHEMBL14671793 | C34H44N6O | 552.767 | 6 / 1 | 7.1 | No |
| 465142 | 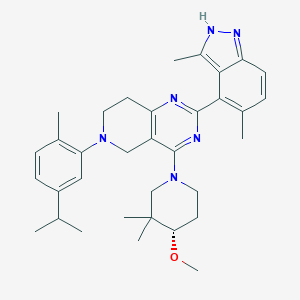 CHEMBL3646936 CHEMBL3646936 | C34H44N6O | 552.767 | 6 / 1 | 7.1 | No |
| 548103 | 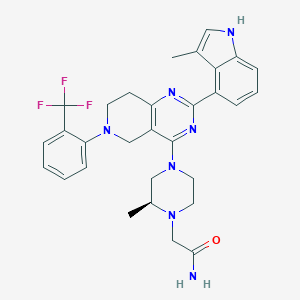 CHEMBL3968102 CHEMBL3968102 | C30H32F3N7O | 563.629 | 9 / 2 | 4.6 | No |
| 557839 | 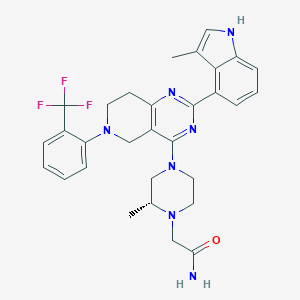 SCHEMBL14662831 SCHEMBL14662831 | C30H32F3N7O | 563.629 | 9 / 2 | 4.6 | No |
| 19546 | 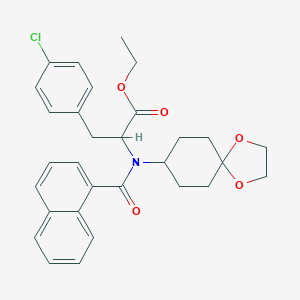 CHEMBL465377 CHEMBL465377 | C30H32ClNO5 | 522.038 | 5 / 0 | 6.3 | No |
| 21413 | 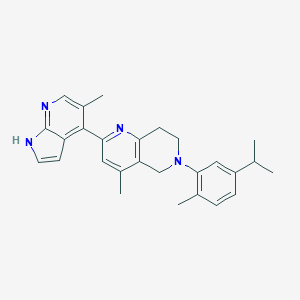 CHEMBL3647040 CHEMBL3647040 | C27H30N4 | 410.565 | 3 / 1 | 5.8 | No |
| 21801 |  CHEMBL86661 CHEMBL86661 | C20H17ClN2O2 | 352.818 | 3 / 1 | 4.5 | Yes |
| 557981 | 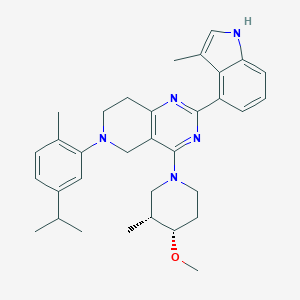 SCHEMBL14671748 SCHEMBL14671748 | C33H41N5O | 523.725 | 5 / 1 | 6.8 | No |
| 465708 | 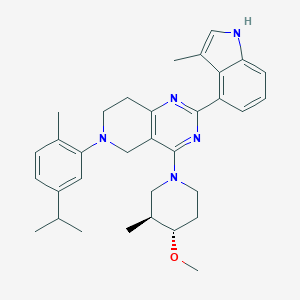 CHEMBL3646919 CHEMBL3646919 | C33H41N5O | 523.725 | 5 / 1 | 6.8 | No |
| 548142 |  CHEMBL3969992 CHEMBL3969992 | C33H41N5O | 523.725 | 5 / 1 | 6.8 | No |
| 23459 | 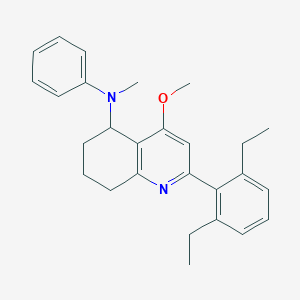 CHEMBL410927 CHEMBL410927 | C27H32N2O | 400.566 | 3 / 0 | 6.5 | No |
| 23962 |  CHEMBL437988 CHEMBL437988 | C46H61ClN8O7 | 873.493 | 8 / 7 | 4.4 | No |
| 24716 | 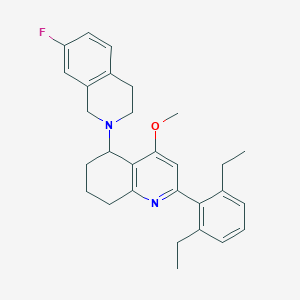 CHEMBL259645 CHEMBL259645 | C29H33FN2O | 444.594 | 4 / 0 | 6.4 | No |
| 558061 | 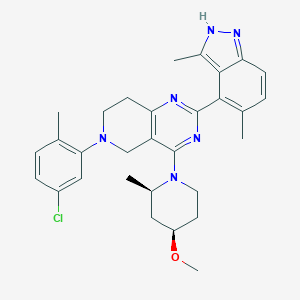 SCHEMBL14663204 SCHEMBL14663204 | C30H35ClN6O | 531.101 | 6 / 1 | 6.2 | No |
| 548170 | 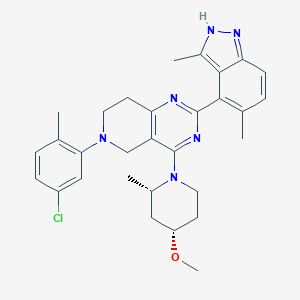 CHEMBL3897947 CHEMBL3897947 | C30H35ClN6O | 531.101 | 6 / 1 | 6.2 | No |
| 459444 | 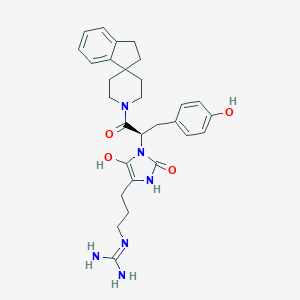 BDBM50290485 BDBM50290485 | C29H36N6O4 | 532.645 | 5 / 5 | 2.2 | No |
| 29782 | 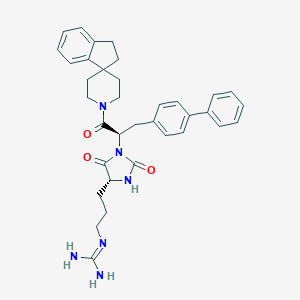 CID 44309234 CID 44309234 | C35H40N6O3 | 592.744 | 4 / 3 | 4.3 | No |
| 29898 | 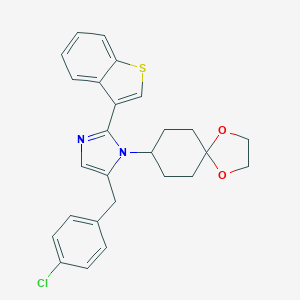 CHEMBL456053 CHEMBL456053 | C26H25ClN2O2S | 465.008 | 4 / 0 | 5.9 | No |
| 30461 | 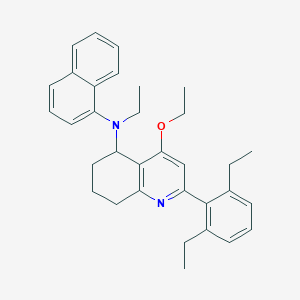 CHEMBL261276 CHEMBL261276 | C33H38N2O | 478.68 | 3 / 0 | 8.5 | No |
| 548221 | 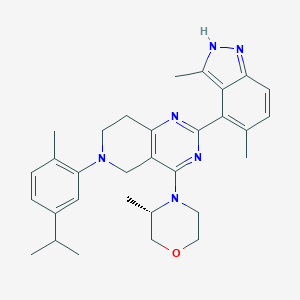 CHEMBL3936892 CHEMBL3936892 | C31H38N6O | 510.686 | 6 / 1 | 6.0 | No |
| 558214 | 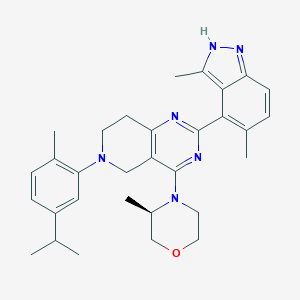 SCHEMBL14663226 SCHEMBL14663226 | C31H38N6O | 510.686 | 6 / 1 | 6.0 | No |
| 31612 | 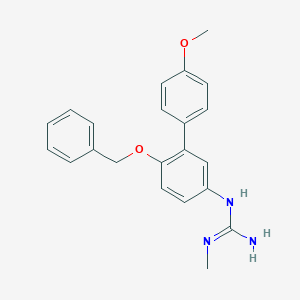 CHEMBL173708 CHEMBL173708 | C22H23N3O2 | 361.445 | 3 / 2 | 3.8 | Yes |
| 558261 | 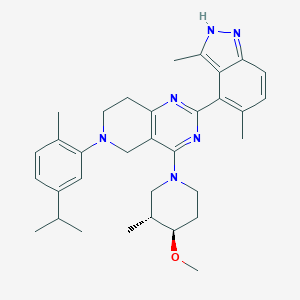 SCHEMBL14671799 SCHEMBL14671799 | C33H42N6O | 538.74 | 6 / 1 | 6.7 | No |
| 548257 | 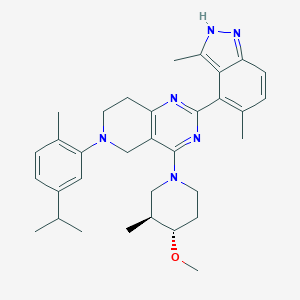 CHEMBL3899595 CHEMBL3899595 | C33H42N6O | 538.74 | 6 / 1 | 6.7 | No |
| 34538 | 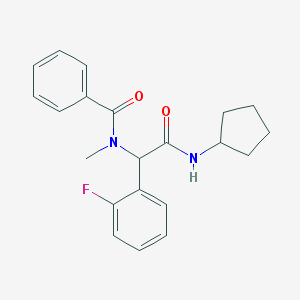 CHEMBL472051 CHEMBL472051 | C21H23FN2O2 | 354.425 | 3 / 1 | 3.8 | Yes |
| 35830 | 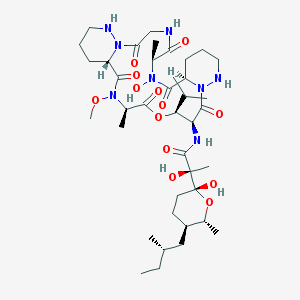 CHEMBL2372061 CHEMBL2372061 | C39H66N8O13 | 855.0 | 15 / 7 | 1.7 | No |
| 37329 | 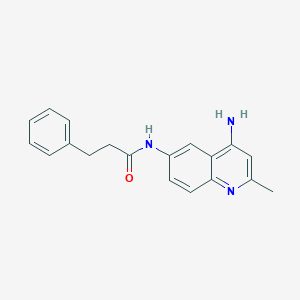 CHEMBL87699 CHEMBL87699 | C19H19N3O | 305.381 | 3 / 2 | 3.0 | Yes |
| 38009 | 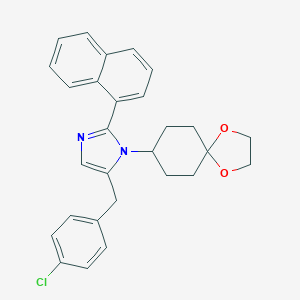 CHEMBL460167 CHEMBL460167 | C28H27ClN2O2 | 458.986 | 3 / 0 | 6.1 | No |
| 38943 | 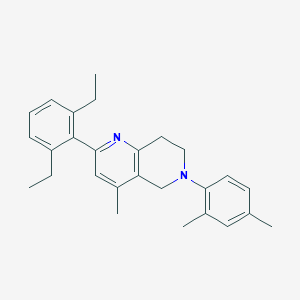 CHEMBL3646988 CHEMBL3646988 | C27H32N2 | 384.567 | 2 / 0 | 6.9 | No |
| 443228 | 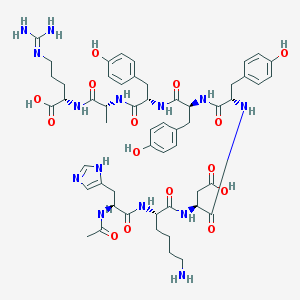 CHEMBL3350664 CHEMBL3350664 | C54H72N14O15 | 1157.25 | 18 / 17 | -3.4 | No |
| 39238 | 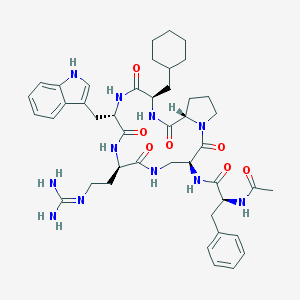 CHEMBL58781 CHEMBL58781 | C44H59N11O7 | 854.026 | 8 / 9 | 2.2 | No |
| 40661 | 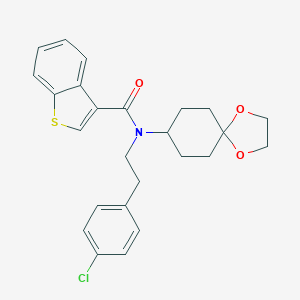 CHEMBL519540 CHEMBL519540 | C25H26ClNO3S | 455.997 | 4 / 0 | 5.7 | No |
| 558534 | 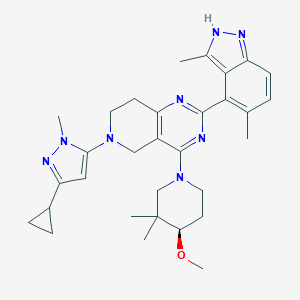 SCHEMBL15105200 SCHEMBL15105200 | C31H40N8O | 540.716 | 7 / 1 | 4.9 | No |
| 548351 |  CHEMBL3895276 CHEMBL3895276 | C31H40N8O | 540.716 | 7 / 1 | 4.9 | No |
| 468038 | 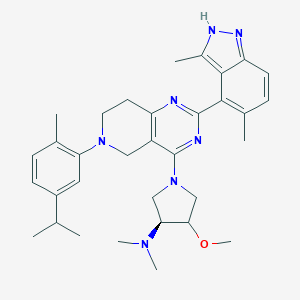 CHEMBL3647020 CHEMBL3647020 | C33H43N7O | 553.755 | 7 / 1 | 5.7 | No |
| 42339 | 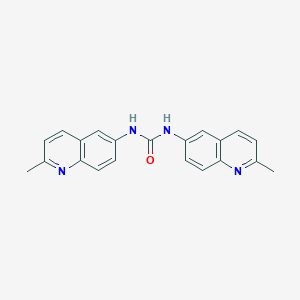 CHEMBL315307 CHEMBL315307 | C21H18N4O | 342.402 | 3 / 2 | 3.8 | Yes |
| 42665 | 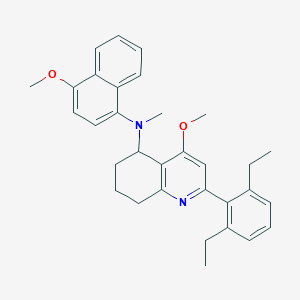 CHEMBL261277 CHEMBL261277 | C32H36N2O2 | 480.652 | 4 / 0 | 7.7 | No |
| 42710 | 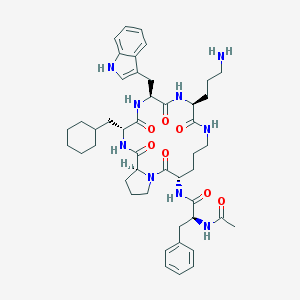 CHEMBL405647 CHEMBL405647 | C46H63N9O7 | 854.066 | 8 / 8 | 3.9 | No |
| 43036 | 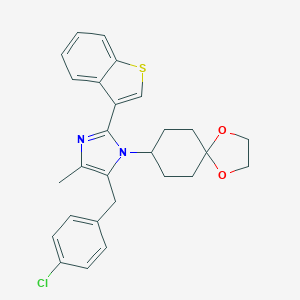 CHEMBL514743 CHEMBL514743 | C27H27ClN2O2S | 479.035 | 4 / 0 | 6.3 | No |
| 43383 | 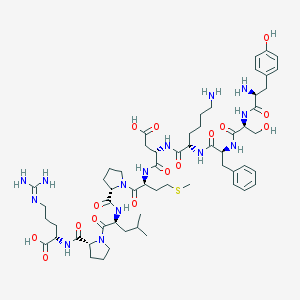 CHEMBL414361 CHEMBL414361 | C58H88N14O15S | 1253.48 | 19 / 15 | -5.5 | No |
| 558631 | 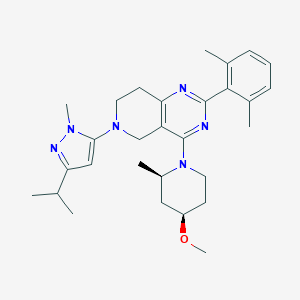 SCHEMBL14660667 SCHEMBL14660667 | C29H40N6O | 488.68 | 6 / 0 | 5.4 | No |
| 548392 | 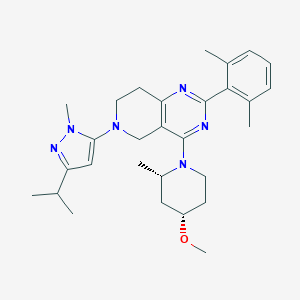 CHEMBL3889744 CHEMBL3889744 | C29H40N6O | 488.68 | 6 / 0 | 5.4 | No |
| 468425 | 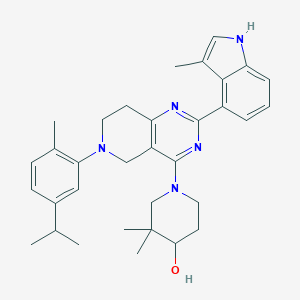 CHEMBL3646911 CHEMBL3646911 | C33H41N5O | 523.725 | 5 / 2 | 6.7 | No |
| 45464 | 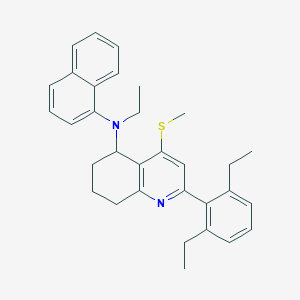 CHEMBL408432 CHEMBL408432 | C32H36N2S | 480.714 | 3 / 0 | 8.7 | No |
| 46819 | 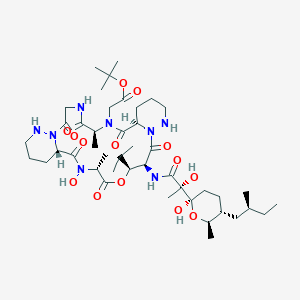 CHEMBL2372064 CHEMBL2372064 | C44H74N8O14 | 939.118 | 16 / 7 | 2.6 | No |
| 47266 | 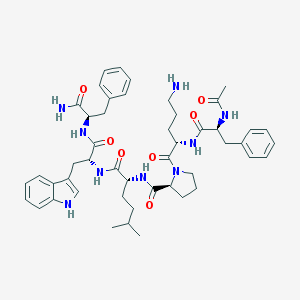 CHEMBL409045 CHEMBL409045 | C48H63N9O7 | 878.088 | 8 / 8 | 3.6 | No |
| 47537 | 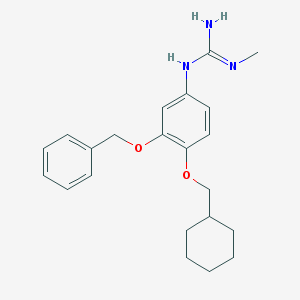 CHEMBL353857 CHEMBL353857 | C22H29N3O2 | 367.493 | 3 / 2 | 4.5 | Yes |
| 48310 | 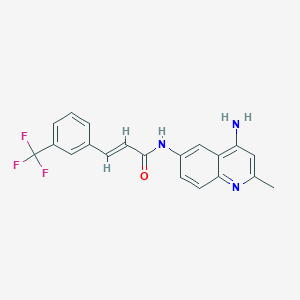 CHEMBL85288 CHEMBL85288 | C20H16F3N3O | 371.363 | 6 / 2 | 4.1 | Yes |
| 49667 | 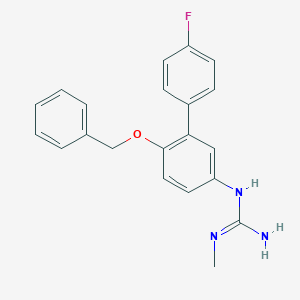 CHEMBL169297 CHEMBL169297 | C21H20FN3O | 349.409 | 3 / 2 | 3.9 | Yes |
| 50063 |  CHEMBL259971 CHEMBL259971 | C32H36N2O | 464.653 | 3 / 0 | 8.1 | No |
| 52076 | 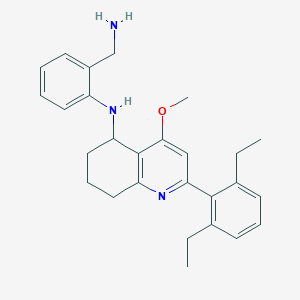 CHEMBL481357 CHEMBL481357 | C27H33N3O | 415.581 | 4 / 2 | 5.8 | No |
| 52317 | 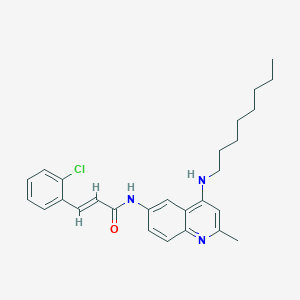 CHEMBL538918 CHEMBL538918 | C27H32ClN3O | 450.023 | 3 / 2 | 7.9 | No |
| 53645 |  CHEMBL1170025 CHEMBL1170025 | C76H123N17O20S2 | 1659.04 | 24 / 21 | 0.3 | No |
| 55517 | 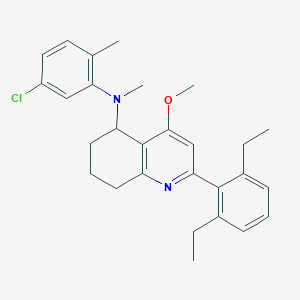 CHEMBL520426 CHEMBL520426 | C28H33ClN2O | 449.035 | 3 / 0 | 7.5 | No |
| 470098 | 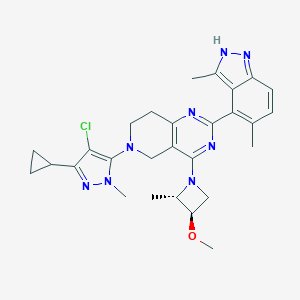 CHEMBL3647026 CHEMBL3647026 | C28H33ClN8O | 533.077 | 7 / 1 | 4.4 | No |
| 58174 | 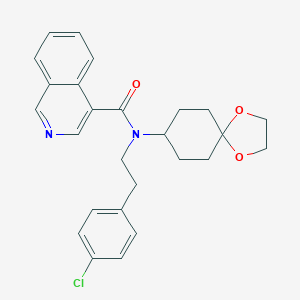 CHEMBL513247 CHEMBL513247 | C26H27ClN2O3 | 450.963 | 4 / 0 | 4.9 | Yes |
| 58302 |  CHEMBL2372087 CHEMBL2372087 | C45H70N8O13 | 931.098 | 15 / 7 | 3.2 | No |
| 58763 |  CID 44309036 CID 44309036 | C33H38N6O3 | 566.706 | 4 / 3 | 3.9 | No |
| 59396 |  CHEMBL82917 CHEMBL82917 | C19H27N3O | 313.445 | 3 / 2 | 4.6 | Yes |
| 59576 |  CHEMBL259642 CHEMBL259642 | C31H34N2O | 450.626 | 3 / 0 | 7.7 | No |
| 61386 |  CHEMBL467571 CHEMBL467571 | C28H37ClN2O3 | 485.065 | 4 / 0 | 5.6 | No |
| 61746 | 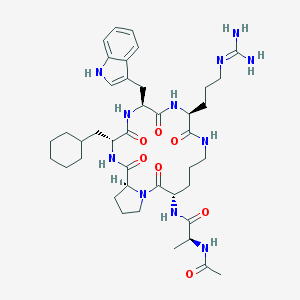 CHEMBL267491 CHEMBL267491 | C41H61N11O7 | 820.009 | 8 / 9 | 1.7 | No |
| 62121 |  CHEMBL409128 CHEMBL409128 | C29H33ClN2O | 461.046 | 3 / 0 | 6.9 | No |
| 62785 | 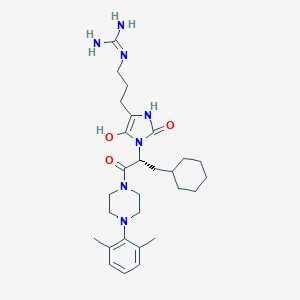 BDBM50290463 BDBM50290463 | C28H43N7O3 | 525.698 | 5 / 4 | 3.2 | No |
| 63632 |  CHEMBL217053 CHEMBL217053 | C18H13BrN2O4 | 401.216 | 5 / 1 | 3.5 | Yes |
| 63692 | 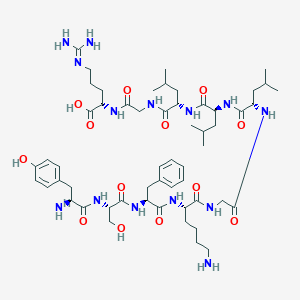 CHEMBL408796 CHEMBL408796 | C55H88N14O13 | 1153.39 | 16 / 16 | -1.8 | No |
| 548643 | 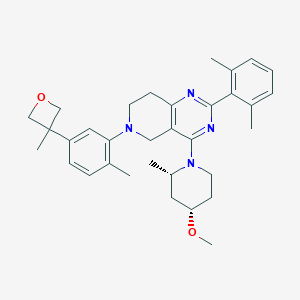 CHEMBL3971727 CHEMBL3971727 | C33H42N4O2 | 526.725 | 6 / 0 | 6.2 | No |
| 559258 | 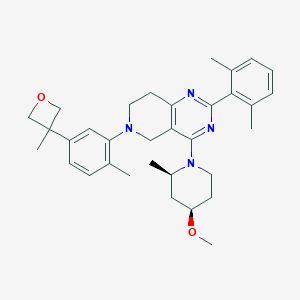 SCHEMBL14660427 SCHEMBL14660427 | C33H42N4O2 | 526.725 | 6 / 0 | 6.2 | No |
| 64356 | 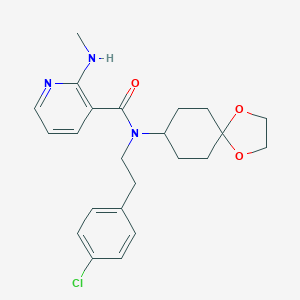 CHEMBL488389 CHEMBL488389 | C23H28ClN3O3 | 429.945 | 5 / 1 | 4.5 | Yes |
| 65043 | 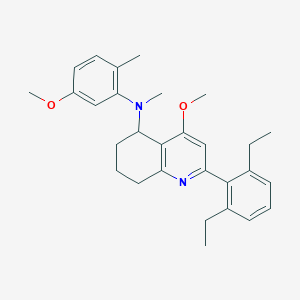 CHEMBL480892 CHEMBL480892 | C29H36N2O2 | 444.619 | 4 / 0 | 6.9 | No |
| 65302 | 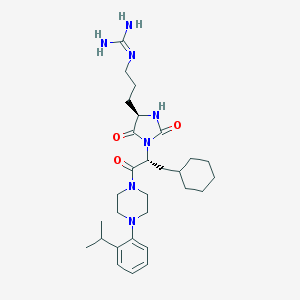 CHEMBL305486 CHEMBL305486 | C29H45N7O3 | 539.725 | 5 / 3 | 3.7 | No |
| 65343 |  CHEMBL2373534 CHEMBL2373534 | C47H69N11O7 | 900.139 | 10 / 10 | 1.2 | No |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218