

You can:
| Name | P2Y purinoceptor 2 |
|---|---|
| Species | Mus musculus (Mouse) |
| Gene | P2ry2 |
| Synonym | Purinergic receptor P2Y2 receptor P2Y2 P2Y purinoceptor 2 ATP receptor [ Show all ] |
| Disease | N/A for non-human GPCRs |
| Length | 373 |
| Amino acid sequence | MAADLEPWNSTINGTWEGDELGYKCRFNEDFKYVLLPVSYGVVCVLGLCLNVVALYIFLCRLKTWNASTTYMFHLAVSDSLYAASLPLLVYYYARGDHWPFSTVLCKLVRFLFYTNLYCSILFLTCISVHRCLGVLRPLHSLRWGRARYARRVAAVVWVLVLACQAPVLYFVTTSVRGTRITCHDTSARELFSHFVAYSSVMLGLLFAVPFSVILVCYVLMARRLLKPAYGTTGGLPRAKRKSVRTIALVLAVFALCFLPFHVTRTLYYSFRSLDLSCHTLNAINMAYKITRPLASANSCLDPVLYFLAGQRLVRFARDAKPPTEPTPSPQARRKLGLHRPNRTVRKDLSVSSDDSRRTESTPAGSETKDIRL |
| UniProt | P35383 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL1075298 |
| IUPHAR | N/A |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 17591 | 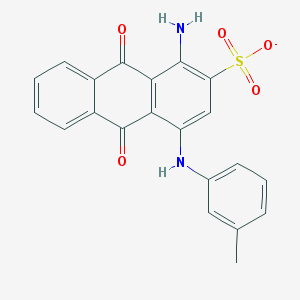 CHEMBL445413 CHEMBL445413 | C21H15N2O5S- | 407.42 | 7 / 2 | 3.9 | Yes |
| 22748 | 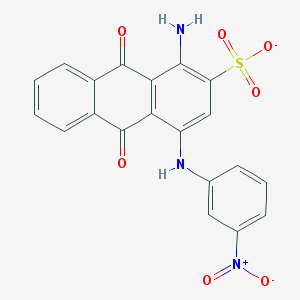 CHEMBL404659 CHEMBL404659 | C20H12N3O7S- | 438.39 | 9 / 2 | 3.3 | Yes |
| 30506 |  CHEMBL271881 CHEMBL271881 | C22H17N2O6S- | 437.446 | 8 / 2 | 3.8 | Yes |
| 32089 | 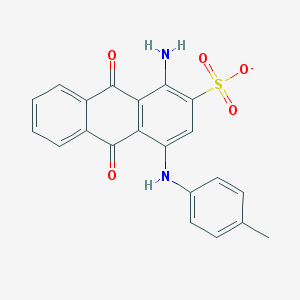 CHEMBL404450 CHEMBL404450 | C21H15N2O5S- | 407.42 | 7 / 2 | 3.9 | Yes |
| 43948 | 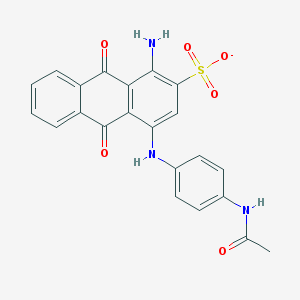 CHEMBL271688 CHEMBL271688 | C22H16N3O6S- | 450.445 | 8 / 3 | 2.7 | Yes |
| 51491 | 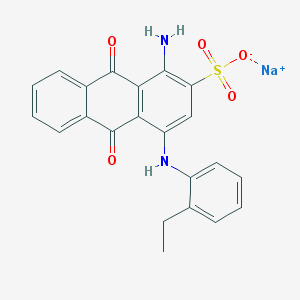 CHEMBL402238 CHEMBL402238 | C22H17N2NaO5S | 444.437 | 7 / 2 | N/A | N/A |
| 53657 | 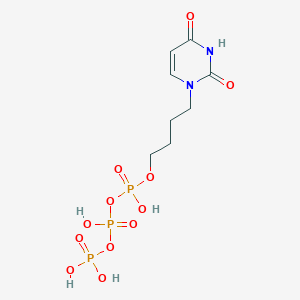 CHEMBL572655 CHEMBL572655 | C8H15N2O12P3 | 424.131 | 12 / 5 | -4.0 | No |
| 58186 | 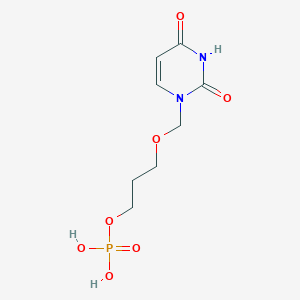 CHEMBL574076 CHEMBL574076 | C8H13N2O7P | 280.173 | 7 / 3 | -2.3 | Yes |
| 68733 | 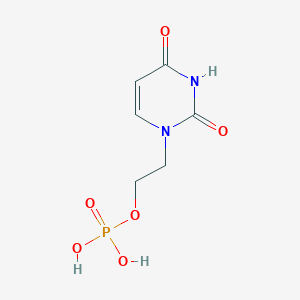 CHEMBL574998 CHEMBL574998 | C6H9N2O6P | 236.12 | 6 / 3 | -2.6 | Yes |
| 70786 | 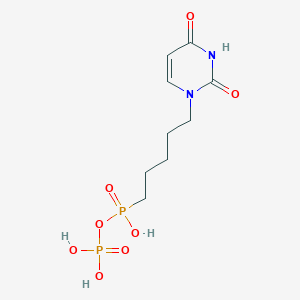 CHEMBL574529 CHEMBL574529 | C9H16N2O8P2 | 342.181 | 8 / 4 | -2.6 | Yes |
| 88042 |  CHEMBL582936 CHEMBL582936 | C7H13N2O11P3 | 394.105 | 11 / 5 | -4.5 | No |
| 90541 | 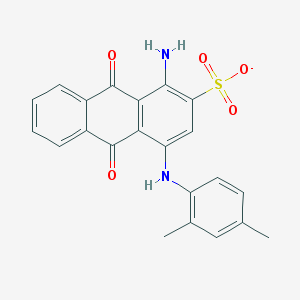 CHEMBL256688 CHEMBL256688 | C22H17N2O5S- | 421.447 | 7 / 2 | 4.2 | Yes |
| 92431 | 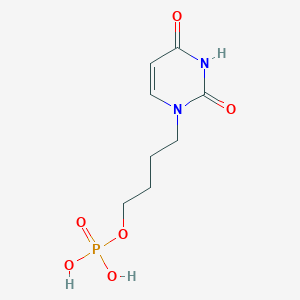 CHEMBL574987 CHEMBL574987 | C8H13N2O6P | 264.174 | 6 / 3 | -1.9 | Yes |
| 93851 | 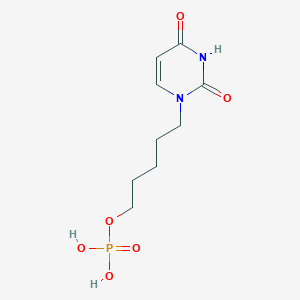 CHEMBL572422 CHEMBL572422 | C9H15N2O6P | 278.201 | 6 / 3 | -1.5 | Yes |
| 99353 | 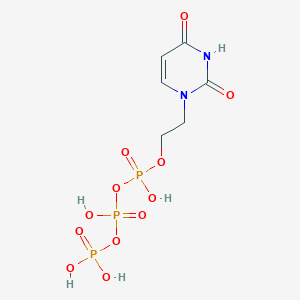 CHEMBL573824 CHEMBL573824 | C6H11N2O12P3 | 396.077 | 12 / 5 | -4.8 | No |
| 108982 | 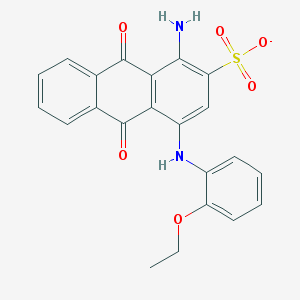 CHEMBL403051 CHEMBL403051 | C22H17N2O6S- | 437.446 | 8 / 2 | 3.8 | Yes |
| 130742 | 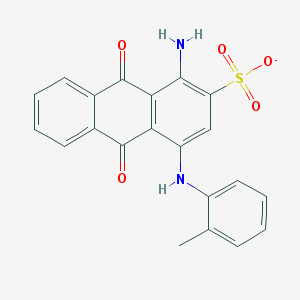 CHEMBL256864 CHEMBL256864 | C21H15N2O5S- | 407.42 | 7 / 2 | 3.9 | Yes |
| 157776 | 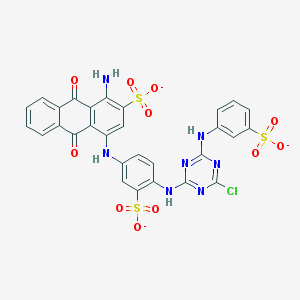 Basilen Blue Basilen Blue | C29H17ClN7O11S3-3 | 771.123 | 18 / 4 | 4.1 | No |
| 159156 |  CHEMBL256689 CHEMBL256689 | C22H17N2O5S- | 421.447 | 7 / 2 | 4.2 | Yes |
| 159956 |  Acyclo-UTP Acyclo-UTP | C7H13N2O13P3 | 426.103 | 13 / 5 | -4.8 | No |
| 160499 | 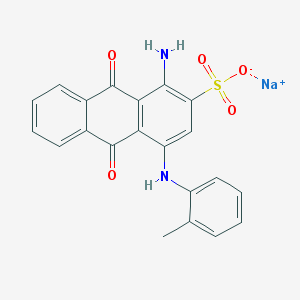 25492-67-5 25492-67-5 | C21H15N2NaO5S | 430.41 | 7 / 2 | N/A | N/A |
| 160747 | 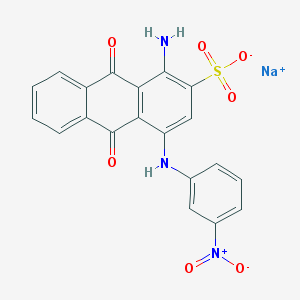 CHEMBL404659 CHEMBL404659 | C20H12N3NaO7S | 461.38 | 9 / 2 | N/A | N/A |
| 169727 | 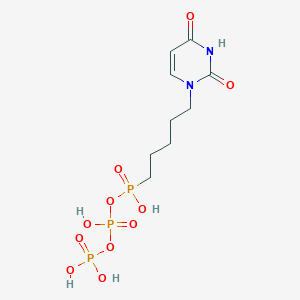 CHEMBL584184 CHEMBL584184 | C9H17N2O11P3 | 422.159 | 11 / 5 | -3.7 | No |
| 169935 | 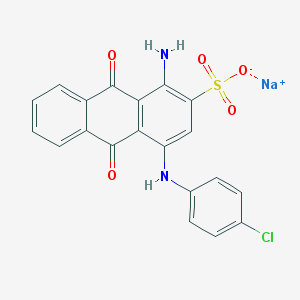 PSB 069 PSB 069 | C20H12ClN2NaO5S | 450.825 | 7 / 2 | N/A | N/A |
| 171594 | 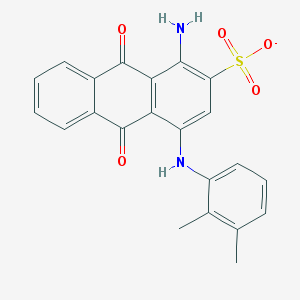 CHEMBL402239 CHEMBL402239 | C22H17N2O5S- | 421.447 | 7 / 2 | 4.2 | Yes |
| 177470 | 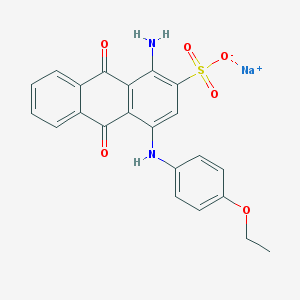 CHEMBL271881 CHEMBL271881 | C22H17N2NaO6S | 460.436 | 8 / 2 | N/A | N/A |
| 188336 | 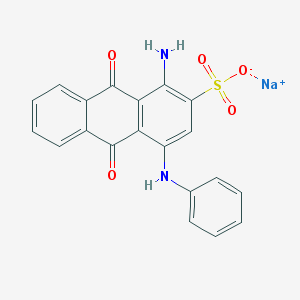 Acid Blue 25 Acid Blue 25 | C20H13N2NaO5S | 416.383 | 7 / 2 | N/A | N/A |
| 199361 | 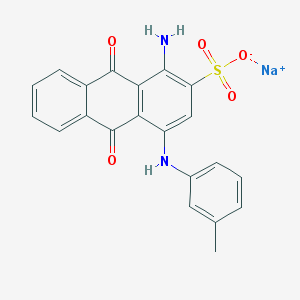 CHEMBL445413 CHEMBL445413 | C21H15N2NaO5S | 430.41 | 7 / 2 | N/A | N/A |
| 205779 | 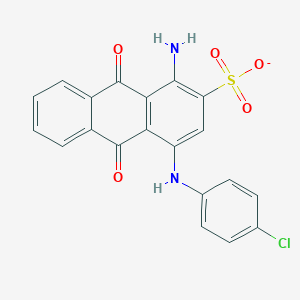 BDBM50227019 BDBM50227019 | C20H12ClN2O5S- | 427.835 | 7 / 2 | 4.1 | Yes |
| 207878 | 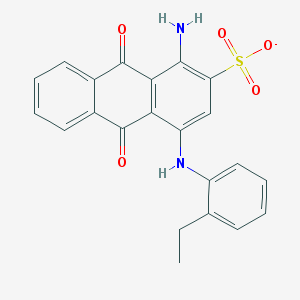 CHEMBL402238 CHEMBL402238 | C22H17N2O5S- | 421.447 | 7 / 2 | 4.3 | Yes |
| 214609 |  CHEMBL401735 CHEMBL401735 | C21H15N2O6S- | 423.419 | 8 / 2 | 3.5 | Yes |
| 220380 | 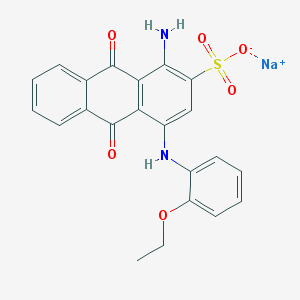 CHEMBL403051 CHEMBL403051 | C22H17N2NaO6S | 460.436 | 8 / 2 | N/A | N/A |
| 231467 | 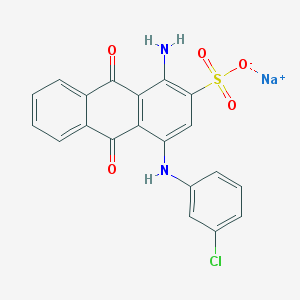 CHEMBL271672 CHEMBL271672 | C20H12ClN2NaO5S | 450.825 | 7 / 2 | N/A | N/A |
| 231761 | 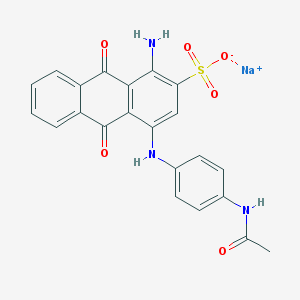 acid blue 40 acid blue 40 | C22H16N3NaO6S | 473.435 | 8 / 3 | N/A | N/A |
| 232079 | 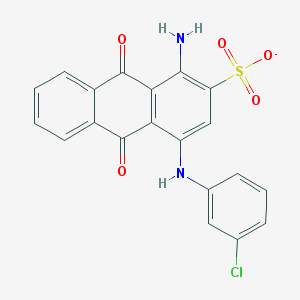 CHEMBL271672 CHEMBL271672 | C20H12ClN2O5S- | 427.835 | 7 / 2 | 4.1 | Yes |
| 246853 | 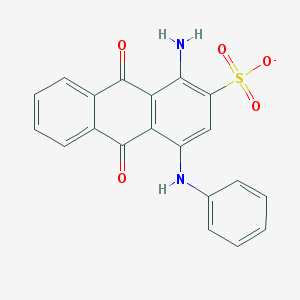 CHEMBL256057 CHEMBL256057 | C20H13N2O5S- | 393.393 | 7 / 2 | 3.5 | Yes |
| 250906 |  CHEMBL605786 CHEMBL605786 | C11H19N2O15P3 | 512.193 | 15 / 7 | -5.3 | No |
| 250907 | 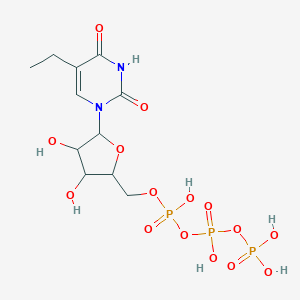 CHEMBL605786 CHEMBL605786 | C11H19N2O15P3 | 512.193 | 15 / 7 | -5.3 | No |
| 254690 | 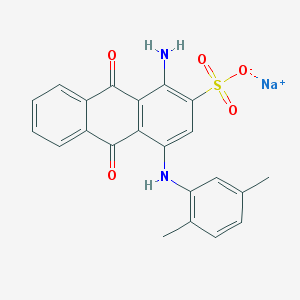 71550-19-1 71550-19-1 | C22H17N2NaO5S | 444.437 | 7 / 2 | N/A | N/A |
| 258881 | 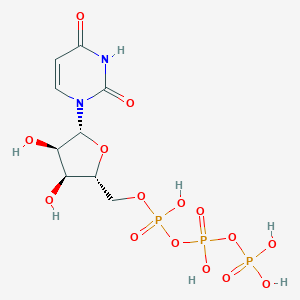 uridine 5'-triphosphate uridine 5'-triphosphate | C9H15N2O15P3 | 484.139 | 15 / 7 | -5.8 | No |
| 266042 | 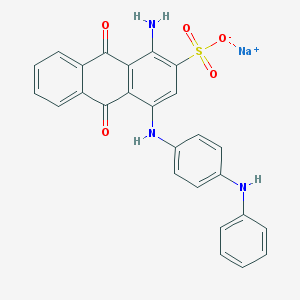 CHEMBL401859 CHEMBL401859 | C26H18N3NaO5S | 507.496 | 8 / 3 | N/A | No |
| 282259 | 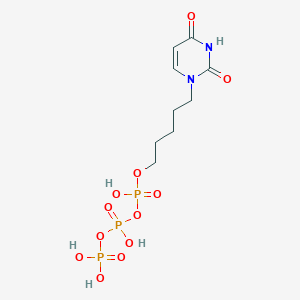 CHEMBL572897 CHEMBL572897 | C9H17N2O12P3 | 438.158 | 12 / 5 | -3.7 | No |
| 284838 | 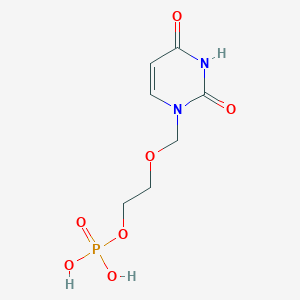 CHEMBL575202 CHEMBL575202 | C7H11N2O7P | 266.146 | 7 / 3 | -2.6 | Yes |
| 290978 | 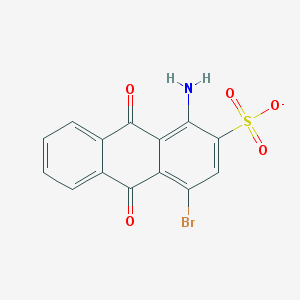 1-amino-4-bromo-9,10-dioxoanthracene-2-sulfonate 1-amino-4-bromo-9,10-dioxoanthracene-2-sulfonate | C14H7BrNO5S- | 381.176 | 6 / 1 | 2.1 | Yes |
| 293876 |  PSB-716 PSB-716 | C21H15N2NaO6S | 446.409 | 8 / 2 | N/A | N/A |
| 296231 | 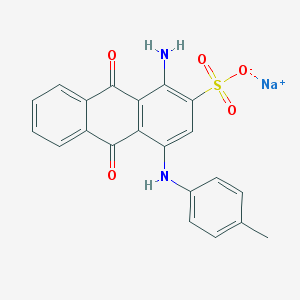 25492-69-7 25492-69-7 | C21H15N2NaO5S | 430.41 | 7 / 2 | N/A | N/A |
| 298329 | 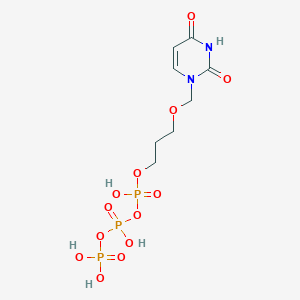 CHEMBL575653 CHEMBL575653 | C8H15N2O13P3 | 440.13 | 13 / 5 | -4.5 | No |
| 316859 | 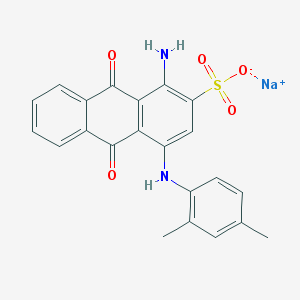 61109-39-5 61109-39-5 | C22H17N2NaO5S | 444.437 | 7 / 2 | N/A | N/A |
| 317274 | 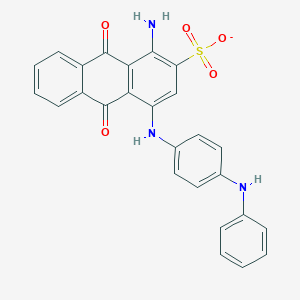 CHEMBL401859 CHEMBL401859 | C26H18N3O5S- | 484.506 | 8 / 3 | 5.0 | Yes |
| 329873 | 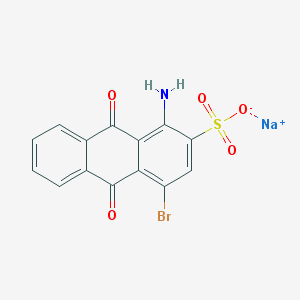 Bromaminic acid sodium salt Bromaminic acid sodium salt | C14H7BrNNaO5S | 404.166 | 6 / 1 | N/A | N/A |
| 339300 | 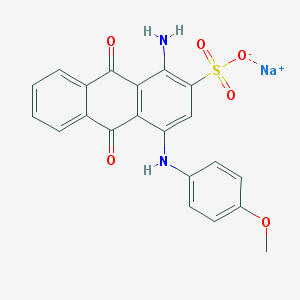 UNII-344PKU9JSY UNII-344PKU9JSY | C21H15N2NaO6S | 446.409 | 8 / 2 | N/A | N/A |
| 362814 |  CHEMBL271689 CHEMBL271689 | C22H15N2O6S- | 435.43 | 8 / 2 | 3.2 | Yes |
| 368115 | 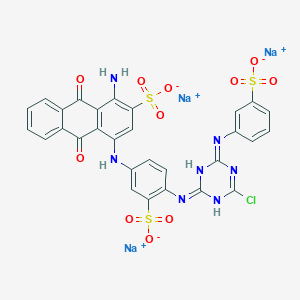 CID 136908877 CID 136908877 | C29H17ClN7Na3O11S3 | 840.092 | 15 / 4 | N/A | No |
| 370183 | 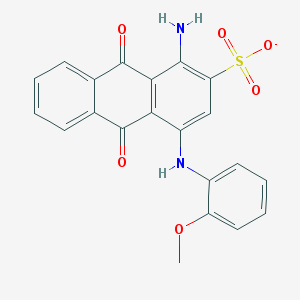 CHEMBL257495 CHEMBL257495 | C21H15N2O6S- | 423.419 | 8 / 2 | 3.5 | Yes |
| 394271 | 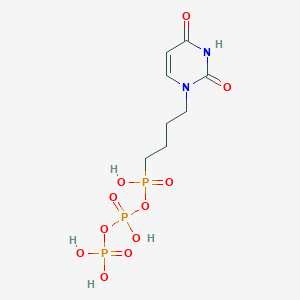 CHEMBL572657 CHEMBL572657 | C8H15N2O11P3 | 408.132 | 11 / 5 | -4.1 | No |
| 426200 | 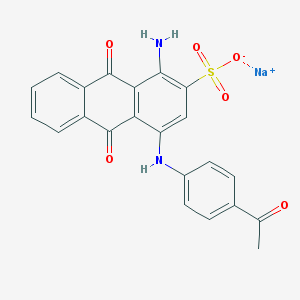 CHEMBL271689 CHEMBL271689 | C22H15N2NaO6S | 458.42 | 8 / 2 | N/A | N/A |
| 432542 | 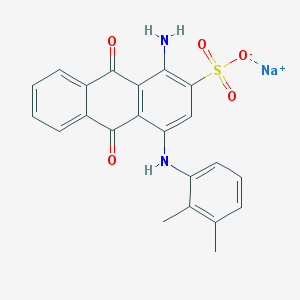 CHEMBL402239 CHEMBL402239 | C22H17N2NaO5S | 444.437 | 7 / 2 | N/A | N/A |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218