

You can:
| Name | CHEMBL1192583 |
|---|---|
| Molecular formula | C9H14N2O2 |
| IUPAC name | 5-(4-methylpiperidin-4-yl)-1,2-oxazol-3-one |
| Molecular weight | 182.223 |
| Hydrogen bond acceptor | 3 |
| Hydrogen bond donor | 2 |
| XlogP | 0.4 |
| Synonyms | CHEMBL543840 5-(4-Methyl-piperidin-4-yl)-isoxazol-3-ol; hydrobromide BDBM50113822 3(2H)-Isoxazolone, 5-(4-methyl-4-piperidinyl)- 439944-41-9 [ Show all ] |
| Inchi Key | JEGDFKOGLTVGEE-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C9H14N2O2/c1-9(2-4-10-5-3-9)7-6-8(12)11-13-7/h6,10H,2-5H2,1H3,(H,11,12) |
| PubChem CID | 11118898 |
| ChEMBL | N/A |
| IUPHAR | N/A |
| BindingDB | 50113822 |
| DrugBank | N/A |
Structure | 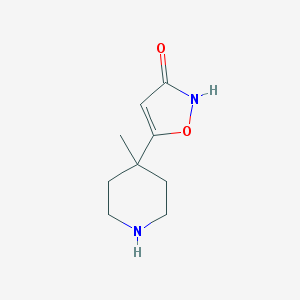 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
You can:
| GLASS ID | Name | UniProt | Gene | Species | Length |
|---|---|---|---|---|---|
| 148807 | Gamma-aminobutyric acid type B receptor subunit 1 | Q9Z0U4 | Gabbr1 | Rattus norvegicus (Rat) | 991 |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218