
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MLRVVEGIFIFVVVSESVFGVLGNGFIGLVNCIDCAKNKLSTIGFILTGLAISRIFLIWIIITDGFIQIFSPNIYASGNLIEYISYFWVIGNQSSMWFATSLSIFYFLKIANFSNYIFLWLKSRTNMVLPFMIVFLLISSLLNFAYIAKILNDYKTKNDTVWDLNMYKSEYFIKQILLNLGVIFFFTLSLITCIFLIISLWRHNRQMQSNVTGLRDSNTEAHVKAMKVLISFIILFILYFIGMAIEISCFTVRENKLLLMFGMTTTAIYPWGHSFILILGNSKLKQASLRVLQQLKCCEKRKNLRVT | |
| CCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCSSSCCCCCHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCSSSSSSCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHCCCCCCCCCCCCC | |
| 9868999999999999999999999999999999967899937899999999999999998689999986411405764455368999998699999999999997078408997489999987258788999999999999999999980563689717985335138999999999999999999999999999999999999851778989999964999999999999999999999999999999837881999999999886399739999918905999999999838250468999998 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MLRVVEGIFIFVVVSESVFGVLGNGFIGLVNCIDCAKNKLSTIGFILTGLAISRIFLIWIIITDGFIQIFSPNIYASGNLIEYISYFWVIGNQSSMWFATSLSIFYFLKIANFSNYIFLWLKSRTNMVLPFMIVFLLISSLLNFAYIAKILNDYKTKNDTVWDLNMYKSEYFIKQILLNLGVIFFFTLSLITCIFLIISLWRHNRQMQSNVTGLRDSNTEAHVKAMKVLISFIILFILYFIGMAIEISCFTVRENKLLLMFGMTTTAIYPWGHSFILILGNSKLKQASLRVLQQLKCCEKRKNLRVT | |
| 7333333311321331233133102300210011024431310110000002001100211333100000003113423101101010122103001000100100000001231110010133031100110132333232222212211333434211123133331220010012333333232113323330210022023202322444412314002200200030221133223223320011133433210110111100010332332424343012002200320201146673658 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCSSSCCCCCHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCSSSSSSCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHCCCCCCCCCCCCC MLRVVEGIFIFVVVSESVFGVLGNGFIGLVNCIDCAKNKLSTIGFILTGLAISRIFLIWIIITDGFIQIFSPNIYASGNLIEYISYFWVIGNQSSMWFATSLSIFYFLKIANFSNYIFLWLKSRTNMVLPFMIVFLLISSLLNFAYIAKILNDYKTKNDTVWDLNMYKSEYFIKQILLNLGVIFFFTLSLITCIFLIISLWRHNRQMQSNVTGLRDSNTEAHVKAMKVLISFIILFILYFIGMAIEISCFTVRENKLLLMFGMTTTAIYPWGHSFILILGNSKLKQASLRVLQQLKCCEKRKNLRVT | |||||||||||||||||||||||||
| 1 | 5nddA | 0.13 | 0.19 | 0.97 | 1.13 | Download | TGKLTTVFLPIVYTIVFVVALPSNGMALWVFLFRT--KKKAPAVIYMANLALADLLSVIWFPLKIAYHIHGNNWIYGEALCNVLIGFFYANMYCSILFLTCLSVQRAWEI-PMGH-----SRKKANIAIGISLAIWLLILLVTIPLYVVKTIFIPALQITTCHDVLLVGDMFNYFLSLAIGVLFPAFLTASAYVLMIRALADLEDNWETLNDNLKVIEKADNAAQVKDALTKMRAAALDAQKATPPKLEDKSPDSPEMKDFRHGFDILVGQIDDALKLANEGKVKEAQAAAEQLKTTRNAYIQKYLE | |||||||||||||||||||
| 2 | 5tgzA | 0.11 | 0.20 | 0.88 | 2.27 | Download | LNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHS-RSLRCRPSYHFIGSLAVADLLGSVIF-VYSFIDFHVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPK----AVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFPHIDKTYLMFWIGVVS---VLLLFIVYAYMYILWKAHSHAPD-----------QARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRS----------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.08 | 0.17 | 0.92 | 2.23 | Download | SSLALAIAITALYSAVCAVGLLGNVLVMFGIVRY--TKMKTATNIYIFNLALADALATST--LPFQSAKYLMETWFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKA----LDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSWYWDTVTKICVFLFVVPILIITVCYGLMLLRLRSV-----RLLSGSK-EKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG---------- | |||||||||||||||||||
| 4 | 4djh | 0.10 | 0.23 | 0.93 | 1.54 | Download | -SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRY--TKMKTATNIYIFNLALADALVTTT-MPFQSTVYLMNSWPF-GDVLCIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVK----ALDFRTPLKAKIINICIWLLSSVGISAIVLGGKVRDVVECSQFPDDDYSWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNIFTPAYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------- | |||||||||||||||||||
| 5 | 5glh | 0.13 | 0.17 | 0.88 | 1.17 | Download | IKETFKYINTVVSCLVFVLGIIGNSTLLYIIYK-------NGPNILIASLALGDLLHIVIAIPINVYKLLAEDWPFGAEMCKLVPFIQKASVGITVLSLCALSIDRYRAVAS---W----------WTAVEIVLIWVVSVVLA-VPEAIGFDIITMSYLRICLLHPKTAFMQFYWWLFSFYFCLPLAITAFFYTLMTCEMLRKNEGLRLTWDAYLNDHLKQRREVAKTVFCLVLVFALCWLPLHLARILKTLYNNLVLDYIGINMASLNSCANPIALYLVSKRFKNAFKSAL--------------- | |||||||||||||||||||
| 6 | 4djhA | 0.08 | 0.19 | 0.91 | 1.52 | Download | -SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRY--TKMKTATNIYIFNLALADALVTTTMPFQSTVYLMN-SWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFPYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSD-----------RNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------- | |||||||||||||||||||
| 7 | 3uon | 0.10 | 0.23 | 0.92 | 1.68 | Download | -----VVFIVLVAGSLSLVTIIGNILVMVSIKV--NRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVGVRTVED-GECYIQFF-SNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFGGAAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAP-CIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM-------------- | |||||||||||||||||||
| 8 | 4n6hA | 0.08 | 0.17 | 0.92 | 2.56 | Download | --GALAIAITALYSAVCAVGLLGNVLVMFGIVRY--TKMKTATNIYIFNLALADALATST-LPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVK----ALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSV------RLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKCG----------- | |||||||||||||||||||
| 9 | 4djhA | 0.11 | 0.19 | 0.89 | 1.44 | Download | -SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRY--TKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPF--DVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCPVKALDFRTPLKA----KIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLL------------DRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------- | |||||||||||||||||||
| 10 | 1l9hA | 0.11 | 0.21 | 0.94 | 1.75 | Download | AEPWQFSMLAAYMFLLIMLGFPINFLTLYVTVQH--KKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLGGEIALWSLVVLAIERYVVVCPMSNFRFG-----ENHAIMGVAFTWVMALACAAPPLVGWSRYIPEGMQCSCGIDYYTPNESFVIYMFVVHFIIPLIVIFFCYGQLVF----------TVKEAAAATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGPIFMTIPAFFAKTSAVYNPVIYIMMNKQFRNCMVTTLCCGKNPLGDSTTVSK | |||||||||||||||||||
| ||||||||||||||||||||||||||
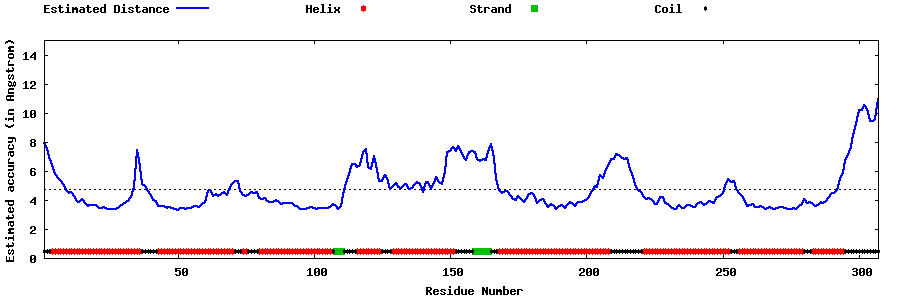
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||