

You can:
| Name | G-protein coupled receptor 35 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | GPR35 |
| Synonym | G-protein coupled receptor 3 GPR35 KYNA receptor Kynurenic acid receptor |
| Disease | N/A |
| Length | 309 |
| Amino acid sequence | MNGTYNTCGSSDLTWPPAIKLGFYAYLGVLLVLGLLLNSLALWVFCCRMQQWTETRIYMTNLAVADLCLLCTLPFVLHSLRDTSDTPLCQLSQGIYLTNRYMSISLVTAIAVDRYVAVRHPLRARGLRSPRQAAAVCAVLWVLVIGSLVARWLLGIQEGGFCFRSTRHNFNSMAFPLLGFYLPLAVVVFCSLKVVTALAQRPPTDVGQAEATRKAARMVWANLLVFVVCFLPLHVGLTVRLAVGWNACALLETIRRALYITSKLSDANCCLDAICYYYMAKEFQEASALAVAPSAKAHKSQDSLCVTLA |
| UniProt | Q9HC97 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | Q9HC97 |
| 3D structure model | This predicted structure model is from GPCR-EXP Q9HC97. |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL1293267 |
| IUPHAR | 102 |
| DrugBank | BE0005562 |
| Name | zaprinast |
|---|---|
| Molecular formula | C13H13N5O2 |
| IUPAC name | 5-(2-propoxyphenyl)-2,6-dihydrotriazolo[4,5-d]pyrimidin-7-one |
| Molecular weight | 271.28 |
| Hydrogen bond acceptor | 5 |
| Hydrogen bond donor | 2 |
| XlogP | 1.6 |
| Synonyms | M&B-22948 7H-1,2,3-Triazolo(4,5-d)pyrimidin-7-one, 1,4-dihydro-5-(2-propoxyphenyl)- NCGC00016105-02 AOB5951 NCGC00024894-08 [ Show all ] |
| Inchi Key | REZGGXNDEMKIQB-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C13H13N5O2/c1-2-7-20-9-6-4-3-5-8(9)11-14-12-10(13(19)15-11)16-18-17-12/h3-6H,2,7H2,1H3,(H2,14,15,16,17,18,19) |
| PubChem CID | 135399235 |
| ChEMBL | CHEMBL28079 |
| IUPHAR | 2919 |
| BindingDB | 14363 |
| DrugBank | N/A |
Structure | 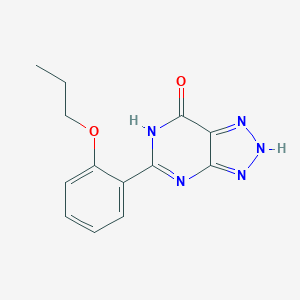 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| EC50 | 110.0 nM | MedChemComm, (2012) 3:10:1270 | ChEMBL |
| EC50 | 130.0 nM | PMID23888932 | BindingDB,ChEMBL |
| EC50 | 140.0 nM | PMID24900508 | BindingDB,ChEMBL |
| EC50 | 160.0 nM | PMID24900447, PMID22572579, PMID21950657 | BindingDB,ChEMBL |
| EC50 | 794.328 nM | PMID16934253 | IUPHAR |
| EC50 | 1960.0 nM | PMID23713606 | BindingDB,ChEMBL |
| EC50 | 2240.0 nM | PMID23713606 | BindingDB,ChEMBL |
| EC50 | 3250.0 nM | MedChemComm, (2012) 3:10:1270 | ChEMBL |
| EC50 | 4200.0 nM | PMID21950657 | BindingDB,ChEMBL |
| EC50 | 5300.0 nM | PMID24900508 | BindingDB,ChEMBL |
| EC50 | 6100.0 nM | PMID24900447 | BindingDB,ChEMBL |
| EC50 | 6200.0 nM | PMID22572579 | BindingDB,ChEMBL |
| Efficacy | 254.0 pM | MedChemComm, (2012) 3:10:1270 | ChEMBL |
| Efficacy | 269.0 picometer | PMID24900447 | ChEMBL |
| IC50 | 1220.0 nM | PMID24900447, PMID21950657 | BindingDB,ChEMBL |
| IC50 | 10500.0 nM | PMID21950657 | BindingDB,ChEMBL |
| Ki | 401.0 nM | PMID23888932 | BindingDB,ChEMBL |
| Ratio | 12.8 - | PMID24900447 | ChEMBL |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218