

You can:
| Name | Thyrotropin receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | TSHR |
| Synonym | LGR3 Thyroid-stimulating hormone receptor Thyrotropin Receptor TSH receptor TSH-R |
| Disease | Thyroid cancer diagnosis Diagnostic test to differentiate primary and secondary hypothyroidism Thyroid cancer |
| Length | 764 |
| Amino acid sequence | MRPADLLQLVLLLDLPRDLGGMGCSSPPCECHQEEDFRVTCKDIQRIPSLPPSTQTLKLIETHLRTIPSHAFSNLPNISRIYVSIDVTLQQLESHSFYNLSKVTHIEIRNTRNLTYIDPDALKELPLLKFLGIFNTGLKMFPDLTKVYSTDIFFILEITDNPYMTSIPVNAFQGLCNETLTLKLYNNGFTSVQGYAFNGTKLDAVYLNKNKYLTVIDKDAFGGVYSGPSLLDVSQTSVTALPSKGLEHLKELIARNTWTLKKLPLSLSFLHLTRADLSYPSHCCAFKNQKKIRGILESLMCNESSMQSLRQRKSVNALNSPLHQEYEENLGDSIVGYKEKSKFQDTHNNAHYYVFFEEQEDEIIGFGQELKNPQEETLQAFDSHYDYTICGDSEDMVCTPKSDEFNPCEDIMGYKFLRIVVWFVSLLALLGNVFVLLILLTSHYKLNVPRFLMCNLAFADFCMGMYLLLIASVDLYTHSEYYNHAIDWQTGPGCNTAGFFTVFASELSVYTLTVITLERWYAITFAMRLDRKIRLRHACAIMVGGWVCCFLLALLPLVGISSYAKVSICLPMDTETPLALAYIVFVLTLNIVAFVIVCCCYVKIYITVRNPQYNPGDKDTKIAKRMAVLIFTDFICMAPISFYALSAILNKPLITVSNSKILLVLFYPLNSCANPFLYAIFTKAFQRDVFILLSKFGICKRQAQAYRGQRVPPKNSTDIQVQKVTHDMRQGLHNMEDVYELIENSHLTPKKQGQISEEYMQTVL |
| UniProt | P16473 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | T60606 |
| ChEMBL | CHEMBL1963 |
| IUPHAR | 255 |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 21 | 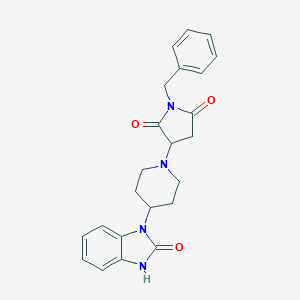 SMR000082495 SMR000082495 | C23H24N4O3 | 404.47 | 4 / 1 | 1.9 | Yes |
| 43 | 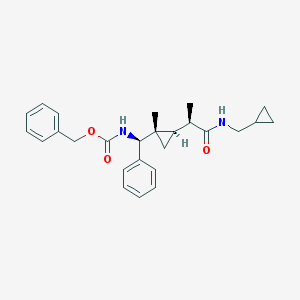 UPCMLD04ASTW002110 UPCMLD04ASTW002110 | C26H32N2O3 | 420.553 | 3 / 2 | 4.6 | Yes |
| 59 |  SMR000006315 SMR000006315 | C18H20ClNO4S | 381.871 | 5 / 1 | 2.8 | Yes |
| 67 | 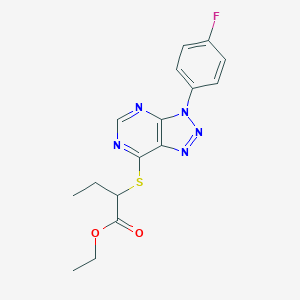 MLS000094053 MLS000094053 | C16H16FN5O2S | 361.395 | 8 / 0 | 3.5 | Yes |
| 109 | 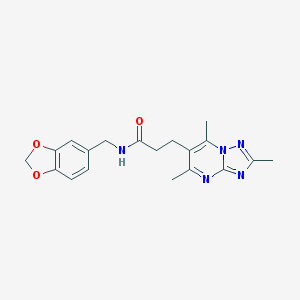 N-(1,3-benzodioxol-5-ylmethyl)-3-(2,5,7-trimethyl[1,2,4]triazolo[1,5-a]pyrimidin-6-yl)propanamide N-(1,3-benzodioxol-5-ylmethyl)-3-(2,5,7-trimethyl[1,2,4]triazolo[1,5-a]pyrimidin-6-yl)propanamide | C19H21N5O3 | 367.409 | 6 / 1 | 2.1 | Yes |
| 161 | 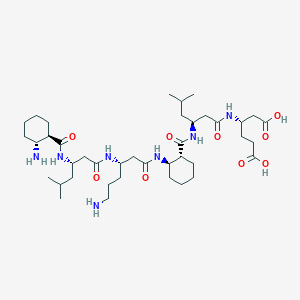 uwisc-betapep-72 uwisc-betapep-72 | C40H71N7O9 | 794.048 | 11 / 9 | -2.2 | No |
| 178 | 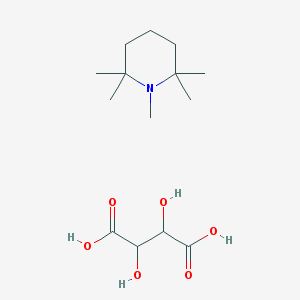 SR-01000838868 SR-01000838868 | C14H27NO6 | 305.371 | 7 / 4 | N/A | N/A |
| 182 | 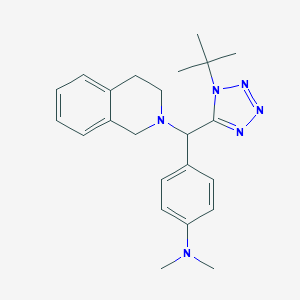 MLS000027156 MLS000027156 | C23H30N6 | 390.535 | 5 / 0 | 3.7 | Yes |
| 203 | 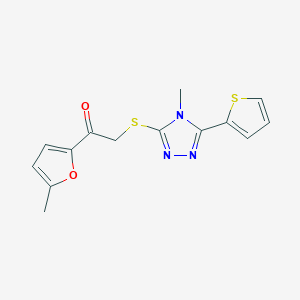 MLS000115918 MLS000115918 | C14H13N3O2S2 | 319.397 | 6 / 0 | 2.9 | Yes |
| 282 |  NCGC00012273 NCGC00012273 | C20H24N6O | 364.453 | 6 / 0 | 1.6 | Yes |
| 354 | 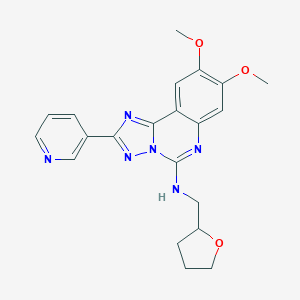 SMR000028904 SMR000028904 | C21H22N6O3 | 406.446 | 8 / 1 | 2.7 | Yes |
| 398 | 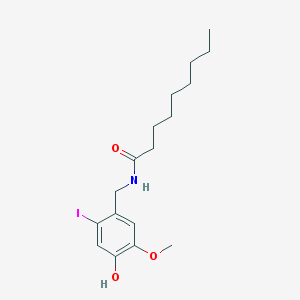 6-Iodonordihydrocapsaicin 6-Iodonordihydrocapsaicin | C17H26INO3 | 419.303 | 3 / 2 | 4.8 | Yes |
| 423 |  SMR000072988 SMR000072988 | C17H15ClN2O4 | 346.767 | 5 / 1 | 3.8 | Yes |
| 425 | 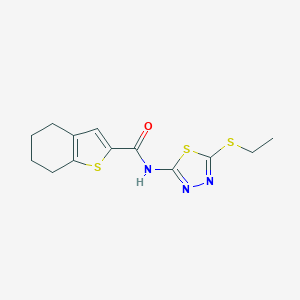 MLS000065772 MLS000065772 | C13H15N3OS3 | 325.463 | 6 / 1 | 4.0 | Yes |
| 493 | 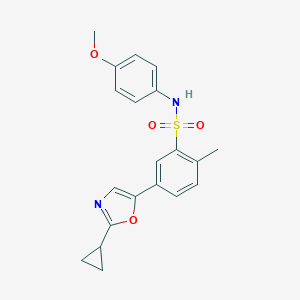 MLS001095734 MLS001095734 | C20H20N2O4S | 384.45 | 6 / 1 | 3.4 | Yes |
| 500 | 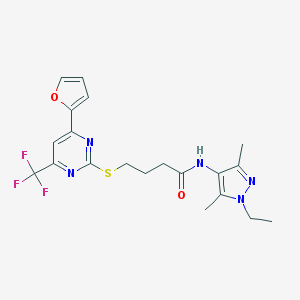 SMR000012485 SMR000012485 | C20H22F3N5O2S | 453.484 | 9 / 1 | 3.3 | Yes |
| 528 | 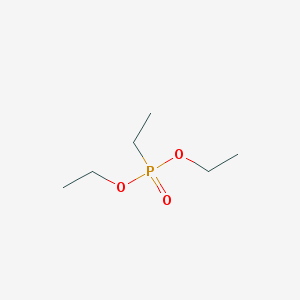 DIETHYL ETHYLPHOSPHONATE DIETHYL ETHYLPHOSPHONATE | C6H15O3P | 166.157 | 3 / 0 | 0.6 | Yes |
| 536 | 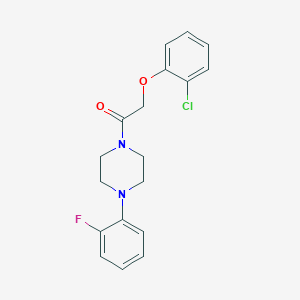 AC1LJP8I AC1LJP8I | C18H18ClFN2O2 | 348.802 | 4 / 0 | 3.7 | Yes |
| 548 | 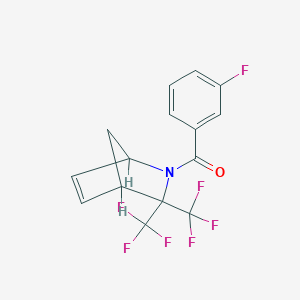 SMR000024980 SMR000024980 | C15H10F7NO | 353.24 | 8 / 0 | 4.2 | Yes |
| 570 |  MLS000113955 MLS000113955 | C11H8BrNO4S | 330.152 | 5 / 1 | 3.6 | Yes |
| 599 | 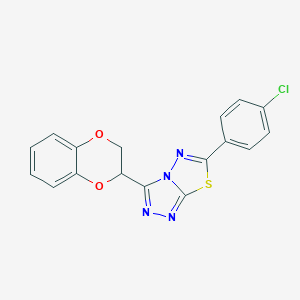 SMR000047551 SMR000047551 | C17H11ClN4O2S | 370.811 | 6 / 0 | 3.8 | Yes |
| 603 |  SMR000004969 SMR000004969 | C26H29ClN6O3 | 509.007 | 7 / 1 | N/A | No |
| 604 | 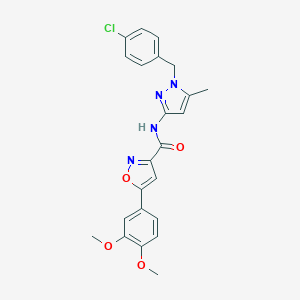 MLS000087136 MLS000087136 | C23H21ClN4O4 | 452.895 | 6 / 1 | 4.4 | Yes |
| 620 | 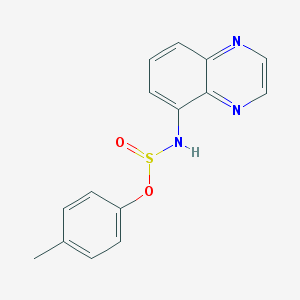 MLS000112244 MLS000112244 | C15H13N3O2S | 299.348 | 6 / 1 | 3.0 | Yes |
| 657 | 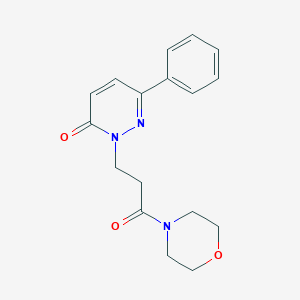 SMR000032427 SMR000032427 | C17H19N3O3 | 313.357 | 4 / 0 | 0.5 | Yes |
| 712 | 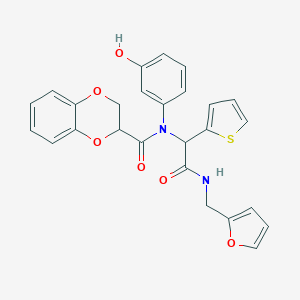 SMR000000828 SMR000000828 | C26H22N2O6S | 490.53 | 7 / 2 | 3.8 | Yes |
| 741 | 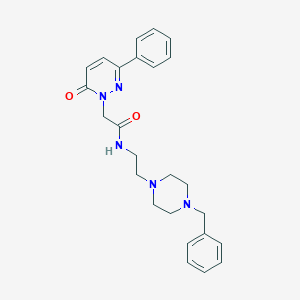 N-[2-(4-benzylpiperazin-1-yl)ethyl]-2-(6-oxo-3-phenylpyridazin-1(6H)-yl)acetamide N-[2-(4-benzylpiperazin-1-yl)ethyl]-2-(6-oxo-3-phenylpyridazin-1(6H)-yl)acetamide | C25H29N5O2 | 431.54 | 5 / 1 | 2.1 | Yes |
| 784 | 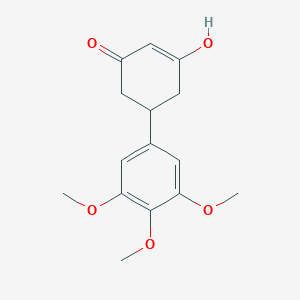 3-hydroxy-5-(3,4,5-trimethoxyphenyl)cyclohex-2-en-1-one 3-hydroxy-5-(3,4,5-trimethoxyphenyl)cyclohex-2-en-1-one | C15H18O5 | 278.304 | 5 / 1 | 1.6 | Yes |
| 874 | 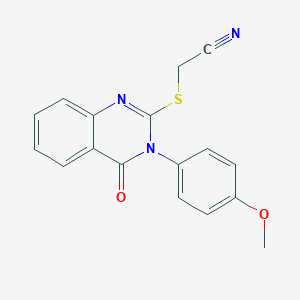 {[3-(4-methoxyphenyl)-4-oxo-3,4-dihydroquinazolin-2-yl]sulfanyl}acetonitrile {[3-(4-methoxyphenyl)-4-oxo-3,4-dihydroquinazolin-2-yl]sulfanyl}acetonitrile | C17H13N3O2S | 323.37 | 5 / 0 | 3.1 | Yes |
| 881 | 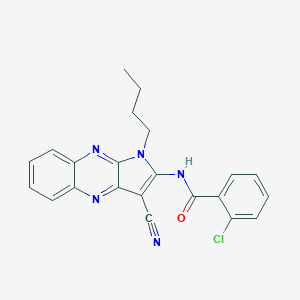 MLS000068755 MLS000068755 | C22H18ClN5O | 403.87 | 4 / 1 | 4.7 | Yes |
| 931 | 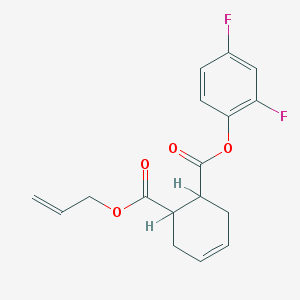 AC1MDXZY AC1MDXZY | C17H16F2O4 | 322.308 | 6 / 0 | 3.5 | Yes |
| 944 | 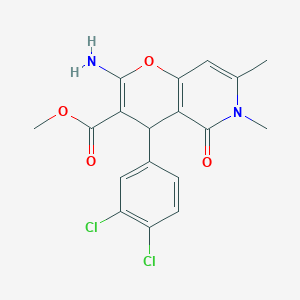 methyl 2-amino-4-(3,4-dichlorophenyl)-6,7-dimethyl-5-oxo-5,6-dihydro-4H-pyrano[3,2-c]pyridine-3-carboxylate methyl 2-amino-4-(3,4-dichlorophenyl)-6,7-dimethyl-5-oxo-5,6-dihydro-4H-pyrano[3,2-c]pyridine-3-carboxylate | C18H16Cl2N2O4 | 395.236 | 5 / 1 | 3.3 | Yes |
| 950 | 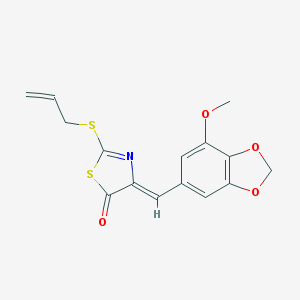 SMR000109040 SMR000109040 | C15H13NO4S2 | 335.392 | 7 / 0 | 3.8 | Yes |
| 954 | 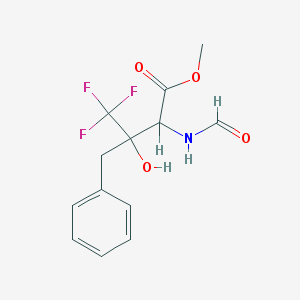 SMR000059935 SMR000059935 | C13H14F3NO4 | 305.253 | 7 / 2 | 1.8 | Yes |
| 955 | 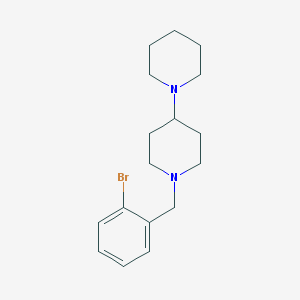 AC1LFMYV AC1LFMYV | C17H25BrN2 | 337.305 | 2 / 0 | 3.9 | Yes |
| 957 |  SMR000062656 SMR000062656 | C19H25N3O2S | 359.488 | 4 / 1 | 3.2 | Yes |
| 958 | 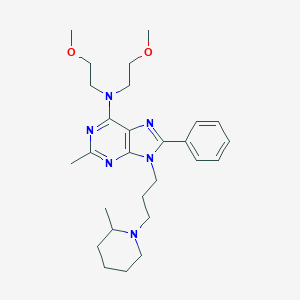 NCGC00090226-01 NCGC00090226-01 | C27H40N6O2 | 480.657 | 7 / 0 | 3.9 | Yes |
| 969 | 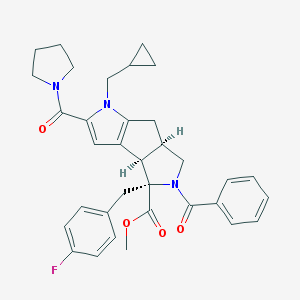 UPCMLD02ASTW001224 UPCMLD02ASTW001224 | C34H36FN3O4 | 569.677 | 5 / 0 | 4.8 | No |
| 1001 | 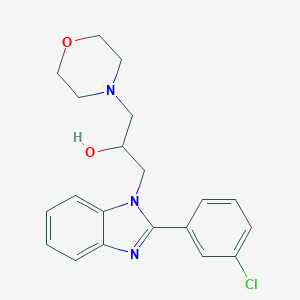 SMR000048110 SMR000048110 | C20H22ClN3O2 | 371.865 | 4 / 1 | 2.7 | Yes |
| 1052 | 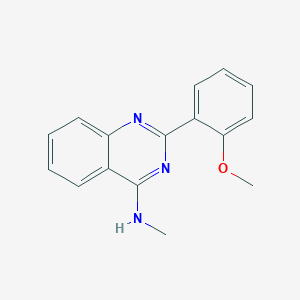 NCGC00012487 NCGC00012487 | C16H15N3O | 265.316 | 4 / 1 | 3.5 | Yes |
| 1069 | 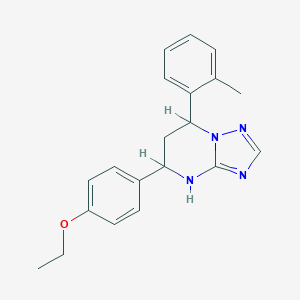 SMR000092165 SMR000092165 | C20H22N4O | 334.423 | 4 / 1 | 4.3 | Yes |
| 1073 | 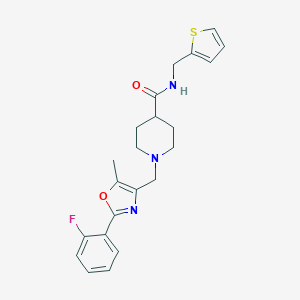 SMR000030768 SMR000030768 | C22H24FN3O2S | 413.511 | 6 / 1 | 3.4 | Yes |
| 1089 |  BAS 09530603 BAS 09530603 | C21H23F2N5O | 399.446 | 6 / 0 | 2.6 | Yes |
| 1102 |  5-(2-Morpholin-4-yl-ethylamino)-2-phenoxymethyl-oxazole-4-carbonitrile 5-(2-Morpholin-4-yl-ethylamino)-2-phenoxymethyl-oxazole-4-carbonitrile | C17H20N4O3 | 328.372 | 7 / 1 | 2.4 | Yes |
| 1113 |  MLS000054921 MLS000054921 | C14H15NO3 | 245.278 | 3 / 0 | 1.3 | Yes |
| 1119 |  AC1LORZK AC1LORZK | C17H19FN2OS | 318.41 | 4 / 0 | 3.9 | Yes |
| 1137 | 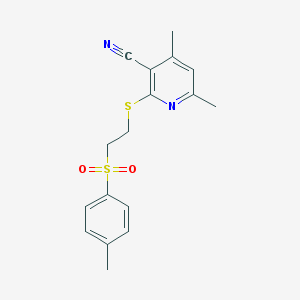 4,6-dimethyl-2-({2-[(4-methylphenyl)sulfonyl]ethyl}sulfanyl)nicotinonitrile 4,6-dimethyl-2-({2-[(4-methylphenyl)sulfonyl]ethyl}sulfanyl)nicotinonitrile | C17H18N2O2S2 | 346.463 | 5 / 0 | 3.5 | Yes |
| 1159 | 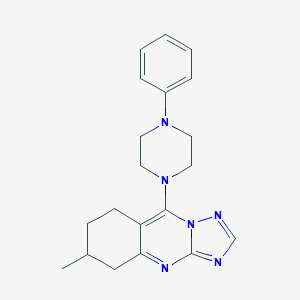 AC1LCTPQ AC1LCTPQ | C20H24N6 | 348.454 | 5 / 0 | 3.8 | Yes |
| 1233 | 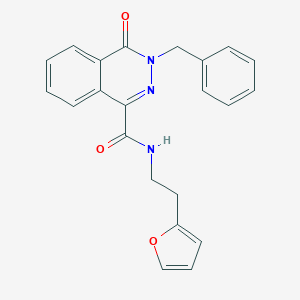 SMR000068198 SMR000068198 | C22H19N3O3 | 373.412 | 4 / 1 | 3.1 | Yes |
| 1243 | 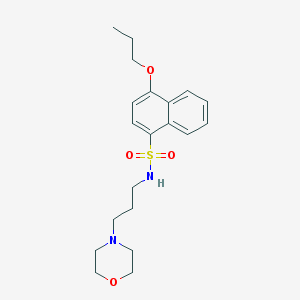 SMR000082223 SMR000082223 | C20H28N2O4S | 392.514 | 6 / 1 | 3.0 | Yes |
| 1272 | 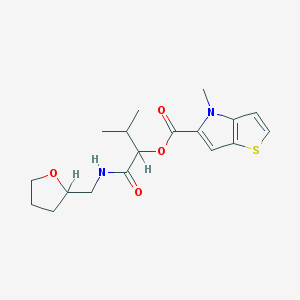 ASN 07460871 ASN 07460871 | C18H24N2O4S | 364.46 | 5 / 1 | 3.1 | Yes |
| 1303 | 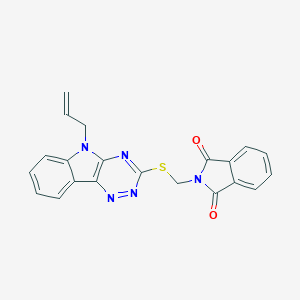 MLS000109191 MLS000109191 | C21H15N5O2S | 401.444 | 6 / 0 | 3.4 | Yes |
| 1324 | 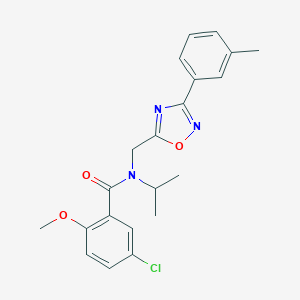 SMR000095165 SMR000095165 | C21H22ClN3O3 | 399.875 | 5 / 0 | 4.6 | Yes |
| 1355 | 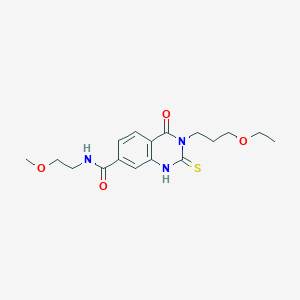 SMR000030857 SMR000030857 | C17H23N3O4S | 365.448 | 5 / 2 | 0.9 | Yes |
| 1393 | 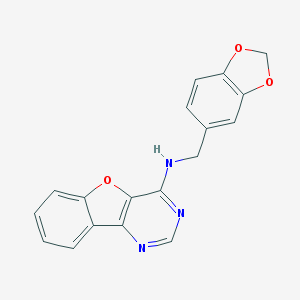 MLS000057790 MLS000057790 | C18H13N3O3 | 319.32 | 6 / 1 | 3.7 | Yes |
| 1442 |  MLS000573697 MLS000573697 | C17H21Cl3N4OS2 | 467.852 | 4 / 3 | 5.6 | No |
| 1505 | 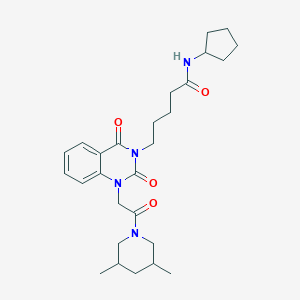 N-cyclopentyl-5-[1-[2-(3,5-dimethylpiperidin-1-yl)-2-oxoethyl]-2,4-dioxo-1,4-dihydroquinazolin-3(2H)-yl]pentanamide N-cyclopentyl-5-[1-[2-(3,5-dimethylpiperidin-1-yl)-2-oxoethyl]-2,4-dioxo-1,4-dihydroquinazolin-3(2H)-yl]pentanamide | C27H38N4O4 | 482.625 | 4 / 1 | 3.3 | Yes |
| 1545 | 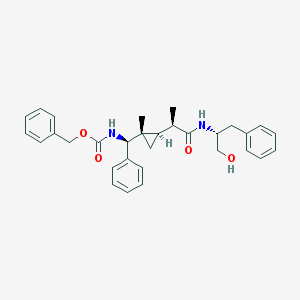 UPCMLD04ADMT001034 UPCMLD04ADMT001034 | C31H36N2O4 | 500.639 | 4 / 3 | 5.2 | No |
| 1554 | 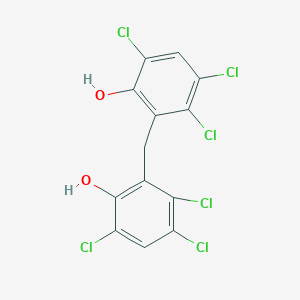 hexachlorophene hexachlorophene | C13H6Cl6O2 | 406.889 | 2 / 2 | 7.5 | No |
| 1586 | 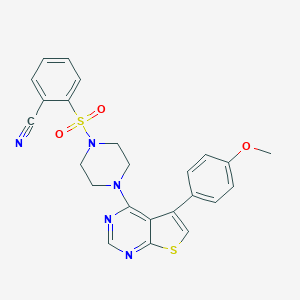 MLS001099132 MLS001099132 | C24H21N5O3S2 | 491.584 | 9 / 0 | 3.9 | Yes |
| 1588 | 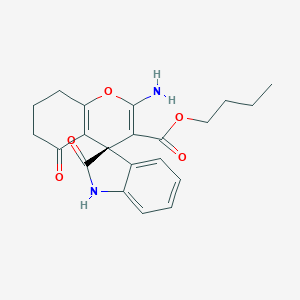 MLS000080402 MLS000080402 | C21H22N2O5 | 382.416 | 6 / 2 | 2.0 | Yes |
| 1601 | 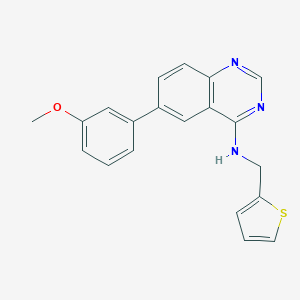 CHEMBL565654 CHEMBL565654 | C20H17N3OS | 347.436 | 5 / 1 | 4.7 | Yes |
| 1610 | 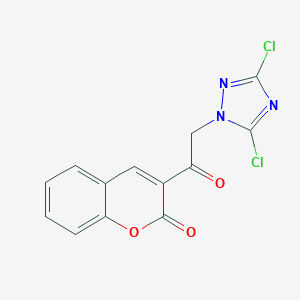 AC1LDFVB AC1LDFVB | C13H7Cl2N3O3 | 324.117 | 5 / 0 | 3.6 | Yes |
| 1614 | 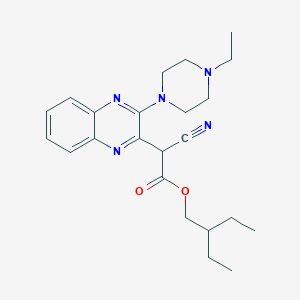 SMR000044012 SMR000044012 | C23H31N5O2 | 409.534 | 7 / 0 | 3.7 | Yes |
| 1618 | 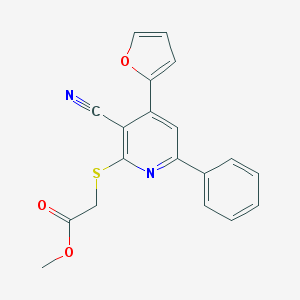 MLS000037786 MLS000037786 | C19H14N2O3S | 350.392 | 6 / 0 | 3.7 | Yes |
| 1622 | 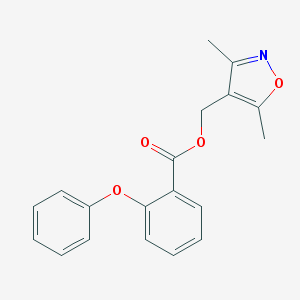 SMR000062829 SMR000062829 | C19H17NO4 | 323.348 | 5 / 0 | 4.0 | Yes |
| 1626 | 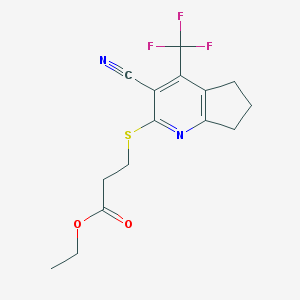 SMR000078129 SMR000078129 | C15H15F3N2O2S | 344.352 | 8 / 0 | 3.2 | Yes |
| 1646 | 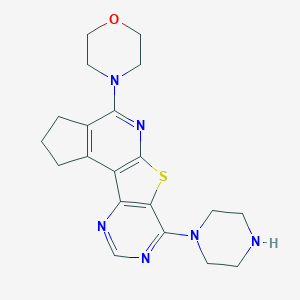 SMR000044353 SMR000044353 | C20H24N6OS | 396.513 | 8 / 1 | 2.6 | Yes |
| 1652 | 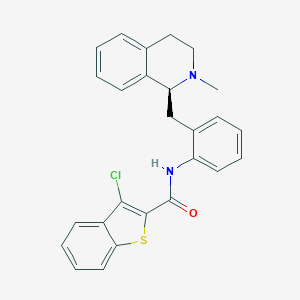 NSC-309874 NSC-309874 | C26H23ClN2OS | 446.993 | 3 / 1 | 6.6 | No |
| 1699 |  Coumarin-3-carboxylic acid Coumarin-3-carboxylic acid | C10H6O4 | 190.154 | 4 / 1 | 2.2 | Yes |
| 1724 | 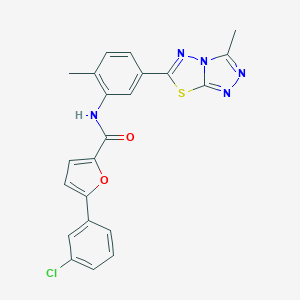 SMR000047763 SMR000047763 | C22H16ClN5O2S | 449.913 | 6 / 1 | 5.2 | No |
| 1778 | 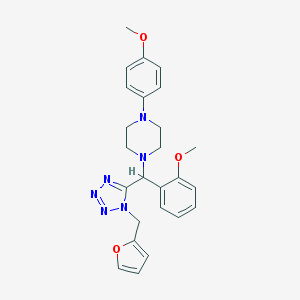 SMR000005020 SMR000005020 | C25H28N6O3 | 460.538 | 8 / 0 | 3.4 | Yes |
| 1803 |  3,4-dichloro-N-(furan-2-ylmethyl)-N-(4-hydroxyphenyl)benzamide 3,4-dichloro-N-(furan-2-ylmethyl)-N-(4-hydroxyphenyl)benzamide | C18H13Cl2NO3 | 362.206 | 3 / 1 | 4.4 | Yes |
| 1818 | 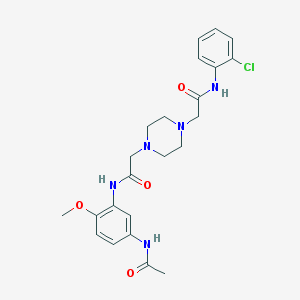 SMR000033485 SMR000033485 | C23H28ClN5O4 | 473.958 | 6 / 3 | 1.7 | Yes |
| 1849 |  UPCMLD04BGHW001051 UPCMLD04BGHW001051 | C19H19NO3 | 309.365 | 3 / 1 | 3.0 | Yes |
| 1850 | 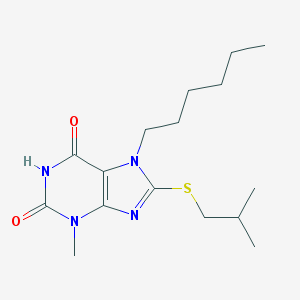 SMR000009624 SMR000009624 | C16H26N4O2S | 338.47 | 4 / 1 | 3.8 | Yes |
| 1961 | 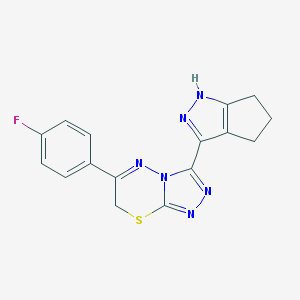 MLS000082992 MLS000082992 | C16H13FN6S | 340.38 | 6 / 1 | 2.7 | Yes |
| 1970 | 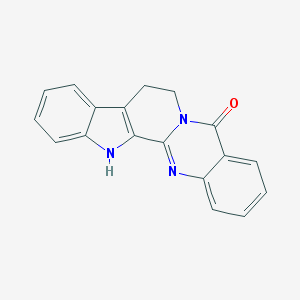 Rutaecarpine Rutaecarpine | C18H13N3O | 287.322 | 2 / 1 | 3.0 | Yes |
| 1974 | 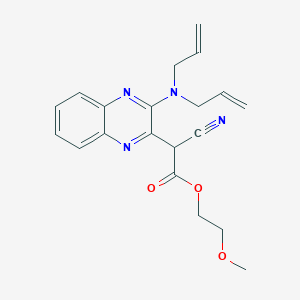 MLS000078748 MLS000078748 | C20H22N4O3 | 366.421 | 7 / 0 | 2.6 | Yes |
| 2018 | 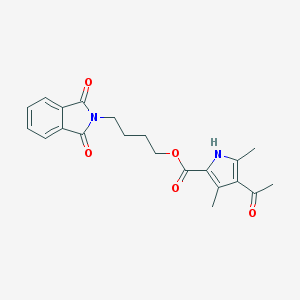 MLS000053810 MLS000053810 | C21H22N2O5 | 382.416 | 5 / 1 | 2.7 | Yes |
| 2036 | 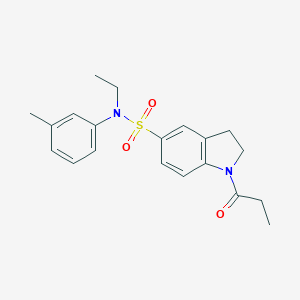 SMR000026069 SMR000026069 | C20H24N2O3S | 372.483 | 4 / 0 | 3.2 | Yes |
| 2066 | 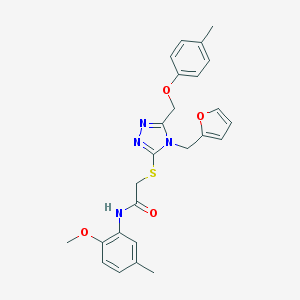 SMR000046468 SMR000046468 | C25H26N4O4S | 478.567 | 7 / 1 | 4.1 | Yes |
| 2077 | 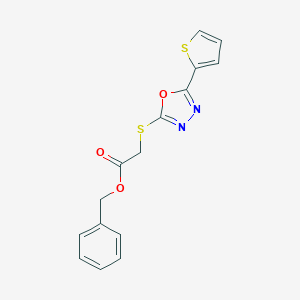 ASN 04370784 ASN 04370784 | C15H12N2O3S2 | 332.392 | 7 / 0 | 3.4 | Yes |
| 2109 | 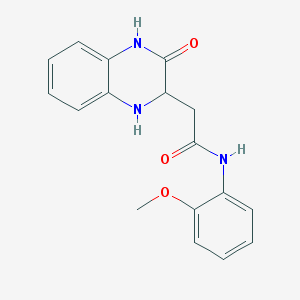 N-(2-methoxyphenyl)-2-(3-oxo-1,2,3,4-tetrahydroquinoxalin-2-yl)acetamide N-(2-methoxyphenyl)-2-(3-oxo-1,2,3,4-tetrahydroquinoxalin-2-yl)acetamide | C17H17N3O3 | 311.341 | 4 / 3 | 1.7 | Yes |
| 2120 |  SMR000106628 SMR000106628 | C16H21N3O3 | 303.362 | 3 / 0 | 0.3 | Yes |
| 2129 | 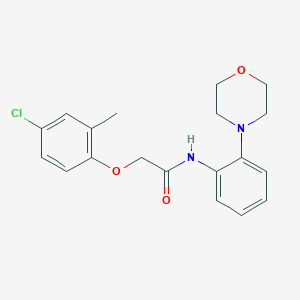 2-(4-chloro-2-methylphenoxy)-N-(2-morpholin-4-ylphenyl)acetamide 2-(4-chloro-2-methylphenoxy)-N-(2-morpholin-4-ylphenyl)acetamide | C19H21ClN2O3 | 360.838 | 4 / 1 | 3.5 | Yes |
| 2135 | 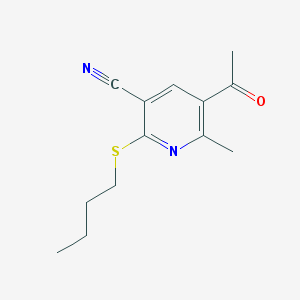 MLS000109037 MLS000109037 | C13H16N2OS | 248.344 | 4 / 0 | 2.8 | Yes |
| 2146 | 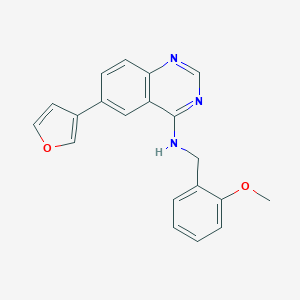 NCGC00012161 NCGC00012161 | C20H17N3O2 | 331.375 | 5 / 1 | 4.0 | Yes |
| 2370 | 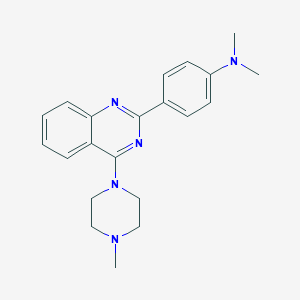 NCGC00012408 NCGC00012408 | C21H25N5 | 347.466 | 5 / 0 | 3.9 | Yes |
| 2385 | 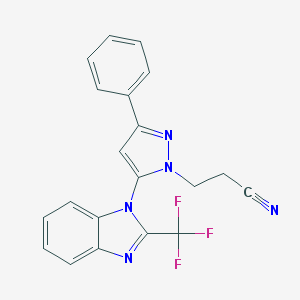 AC1MHBGG AC1MHBGG | C20H14F3N5 | 381.362 | 6 / 0 | 3.9 | Yes |
| 557368 | 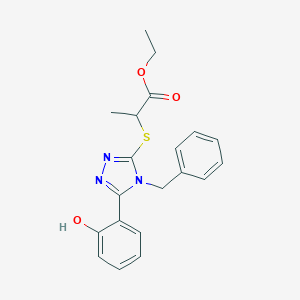 SMR000001967 SMR000001967 | C20H21N3O3S | 383.466 | 6 / 1 | 3.9 | Yes |
| 2913 | 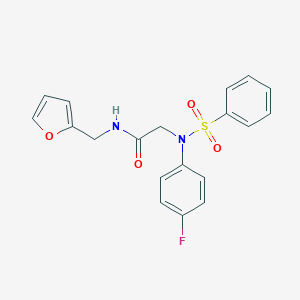 2-[Benzenesulfonyl-(4-fluoro-phenyl)-amino]-N-furan-2-ylmethyl-acetamide 2-[Benzenesulfonyl-(4-fluoro-phenyl)-amino]-N-furan-2-ylmethyl-acetamide | C19H17FN2O4S | 388.413 | 6 / 1 | 2.7 | Yes |
| 3161 | 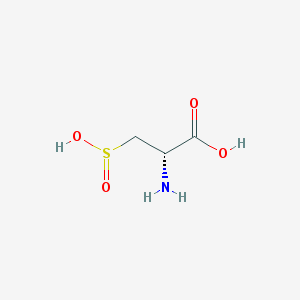 D-Alanine, 3-sulfino- D-Alanine, 3-sulfino- | C3H7NO4S | 153.152 | 6 / 3 | -4.2 | Yes |
| 3168 | 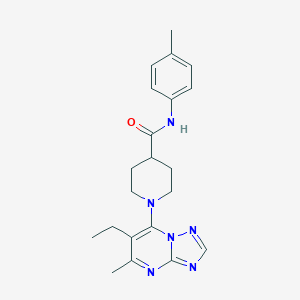 AC1LCKZJ AC1LCKZJ | C21H26N6O | 378.48 | 5 / 1 | 3.4 | Yes |
| 3188 | 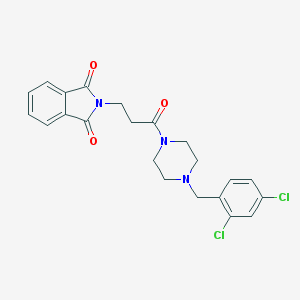 2-{3-[4-(2,4-dichlorobenzyl)piperazin-1-yl]-3-oxopropyl}-1H-isoindole-1,3(2H)-dione 2-{3-[4-(2,4-dichlorobenzyl)piperazin-1-yl]-3-oxopropyl}-1H-isoindole-1,3(2H)-dione | C22H21Cl2N3O3 | 446.328 | 4 / 0 | 2.9 | Yes |
| 3223 | 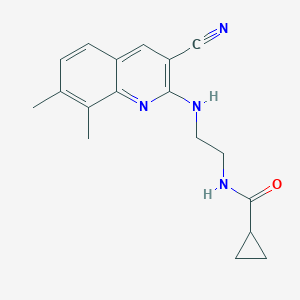 SMR000006848 SMR000006848 | C18H20N4O | 308.385 | 4 / 2 | 3.3 | Yes |
| 3243 | 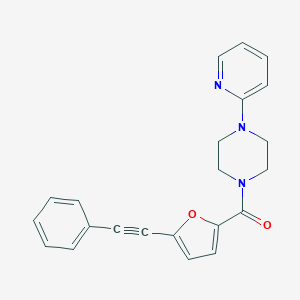 (5-Phenylethynyl-furan-2-yl)-(4-pyridin-2-yl-piperazin-1-yl)-methanone (5-Phenylethynyl-furan-2-yl)-(4-pyridin-2-yl-piperazin-1-yl)-methanone | C22H19N3O2 | 357.413 | 4 / 0 | 3.7 | Yes |
| 3253 |  SMR000000804 SMR000000804 | C21H27N3O3 | 369.465 | 4 / 1 | 3.7 | Yes |
| 3299 | 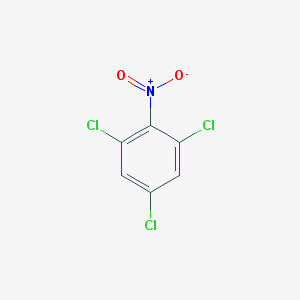 2,4,6-Trichloronitrobenzene 2,4,6-Trichloronitrobenzene | C6H2Cl3NO2 | 226.437 | 2 / 0 | 3.6 | Yes |
| 3312 | 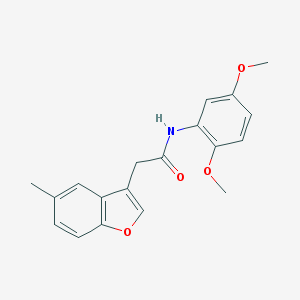 N-(2,5-dimethoxyphenyl)-2-(5-methyl-1-benzofuran-3-yl)acetamide N-(2,5-dimethoxyphenyl)-2-(5-methyl-1-benzofuran-3-yl)acetamide | C19H19NO4 | 325.364 | 4 / 1 | 3.4 | Yes |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218