

You can:
| Name | Metabotropic glutamate receptor 5 |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Grm5 |
| Synonym | glutamate receptor GPRC1E mGlu5 receptor mGluR5 |
| Disease | N/A for non-human GPCRs |
| Length | 1203 |
| Amino acid sequence | MVLLLILSVLLLKEDVRGSAQSSERRVVAHMPGDIIIGALFSVHHQPTVDKVHERKCGAVREQYGIQRVEAMLHTLERINSDPTLLPNITLGCEIRDSCWHSAVALEQSIEFIRDSLISSEEEEGLVRCVDGSSSFRSKKPIVGVIGPGSSSVAIQVQNLLQLFNIPQIAYSATSMDLSDKTLFKYFMRVVPSDAQQARAMVDIVKRYNWTYVSAVHTEGNYGESGMEAFKDMSAKEGICIAHSYKIYSNAGEQSFDKLLKKLRSHLPKARVVACFCEGMTVRGLLMAMRRLGLAGEFLLLGSDGWADRYDVTDGYQREAVGGITIKLQSPDVKWFDDYYLKLRPETNLRNPWFQEFWQHRFQCRLEGFAQENSKYNKTCNSSLTLRTHHVQDSKMGFVINAIYSMAYGLHNMQMSLCPGYAGLCDAMKPIDGRKLLDSLMKTNFTGVSGDMILFDENGDSPGRYEIMNFKEMGKDYFDYINVGSWDNGELKMDDDEVWSKKNNIIRSVCSEPCEKGQIKVIRKGEVSCCWTCTPCKENEYVFDEYTCKACQLGSWPTDDLTGCDLIPVQYLRWGDPEPIAAVVFACLGLLATLFVTVIFIIYRDTPVVKSSSRELCYIILAGICLGYLCTFCLIAKPKQIYCYLQRIGIGLSPAMSYSALVTKTNRIARILAGSKKKICTKKPRFMSACAQLVIAFILICIQLGIIVALFIMEPPDIMHDYPSIREVYLICNTTNLGVVTPLGYNGLLILSCTFYAFKTRNVPANFNEAKYIAFTMYTTCIIWLAFVPIYFGSNYKIITMCFSVSLSATVALGCMFVPKVYIILAKPERNVRSAFTTSTVVRMHVGDGKSSSAASRSSSLVNLWKRRGSSGETLRYKDRRLAQHKSEIECFTPKGSMGNGGRATMSSSNGKSVTWAQNEKSTRGQHLWQRLSVHINKKENPNQTAVIKPFPKSTENRGPGAAAGGGSGPGVAGAGNAGCTATGGPEPPDAGPKALYDVAEAEESFPAAARPRSPSPISTLSHLAGSAGRTDDDAPSLHSETAARSSSSQGSLMEQISSVVTRFTANISELNSMMLSTAATPGPPGTPICSSYLIPKEIQLPTTMTTFAEIQPLPAIEVTGGAQGATGVSPAQETPTGAESAPGKPDLEELVALTPPSPFRDSVDSGSTTPNSPVSESALCIPSSPKYDTLIIRDYTQSSSSL |
| UniProt | P31424 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL2564 |
| IUPHAR | 293 |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 557306 | 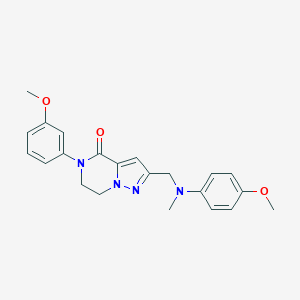 SCHEMBL17334385 SCHEMBL17334385 | C22H24N4O3 | 392.459 | 5 / 0 | 2.9 | Yes |
| 111 | 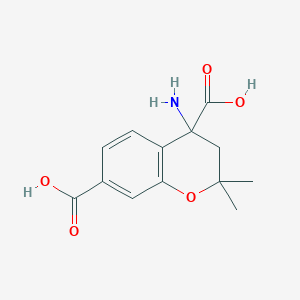 CHEMBL278949 CHEMBL278949 | C13H15NO5 | 265.265 | 6 / 3 | -1.6 | Yes |
| 442 | 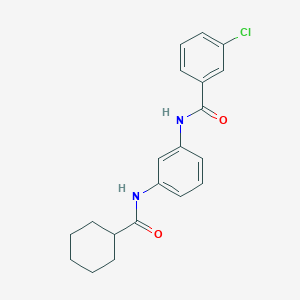 CHEMBL1081848 CHEMBL1081848 | C20H21ClN2O2 | 356.85 | 2 / 2 | 4.6 | Yes |
| 715 | 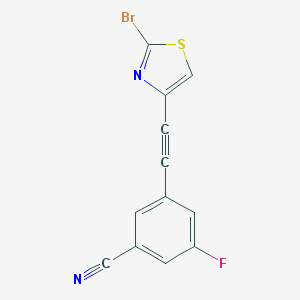 CHEMBL1672280 CHEMBL1672280 | C12H4BrFN2S | 307.14 | 4 / 0 | 3.9 | Yes |
| 1206 | 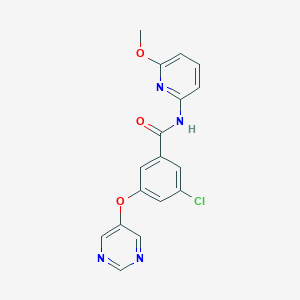 SCHEMBL6918188 SCHEMBL6918188 | C17H13ClN4O3 | 356.766 | 6 / 1 | 2.8 | Yes |
| 1220 |  CHEMBL184047 CHEMBL184047 | C17H11FN6S | 350.375 | 7 / 0 | 3.5 | Yes |
| 1247 | 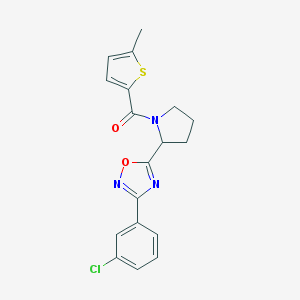 CHEMBL1083341 CHEMBL1083341 | C18H16ClN3O2S | 373.855 | 5 / 0 | 4.4 | Yes |
| 1564 | 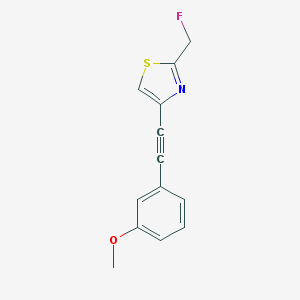 CHEMBL229911 CHEMBL229911 | C13H10FNOS | 247.287 | 4 / 0 | 3.2 | Yes |
| 441736 | 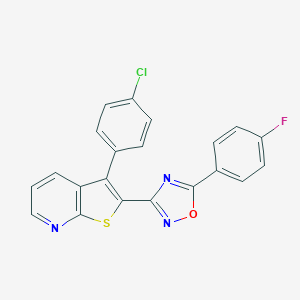 CHEMBL3317731 CHEMBL3317731 | C21H11ClFN3OS | 407.847 | 6 / 0 | 6.1 | No |
| 1755 |  3-iodo-5-(pyridin-2-ylethynyl)benzonitrile 3-iodo-5-(pyridin-2-ylethynyl)benzonitrile | C14H7IN2 | 330.128 | 2 / 0 | 3.3 | Yes |
| 2297 |  CHEMBL2151798 CHEMBL2151798 | C17H26N2O2 | 290.407 | 3 / 1 | 4.0 | Yes |
| 2426 |  CHEMBL239620 CHEMBL239620 | C14H9Cl2N3 | 290.147 | 3 / 0 | 4.1 | Yes |
| 555490 |  AC1N0YGA AC1N0YGA | C3H7NO4S | 153.152 | 5 / 2 | -4.7 | Yes |
| 459256 |  SCHEMBL6928865 SCHEMBL6928865 | C17H11F2N3O2 | 327.291 | 6 / 1 | 2.7 | Yes |
| 441859 | 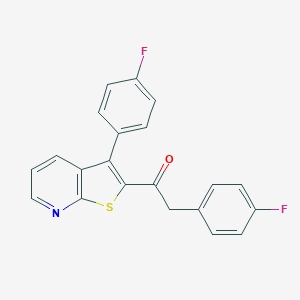 CHEMBL3317726 CHEMBL3317726 | C21H13F2NOS | 365.398 | 5 / 0 | 5.6 | No |
| 557395 | 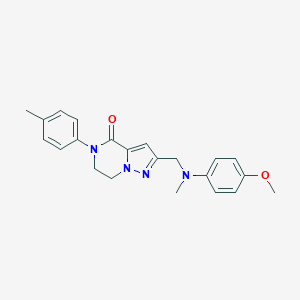 SCHEMBL17334442 SCHEMBL17334442 | C22H24N4O2 | 376.46 | 4 / 0 | 3.3 | Yes |
| 4017 | 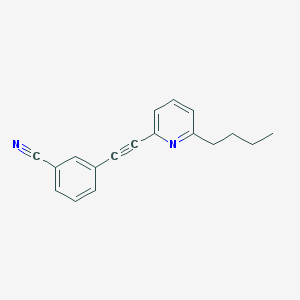 CHEMBL2089180 CHEMBL2089180 | C18H16N2 | 260.34 | 2 / 0 | 4.4 | Yes |
| 4020 | 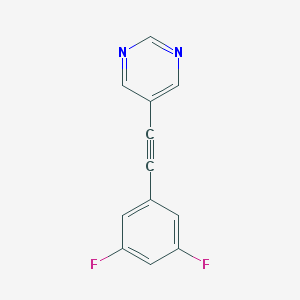 CHEMBL488233 CHEMBL488233 | C12H6F2N2 | 216.191 | 4 / 0 | 2.4 | Yes |
| 4046 | 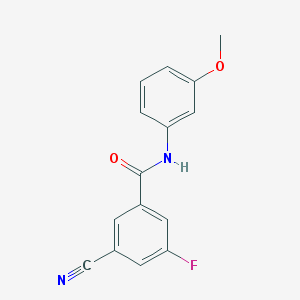 CHEMBL1209470 CHEMBL1209470 | C15H11FN2O2 | 270.263 | 4 / 1 | 2.8 | Yes |
| 557412 | 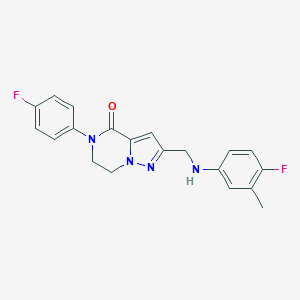 SCHEMBL17334231 SCHEMBL17334231 | C20H18F2N4O | 368.388 | 5 / 1 | 3.4 | Yes |
| 463456 | 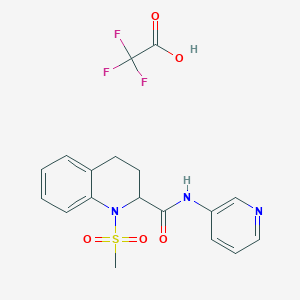 CHEMBL3182872 CHEMBL3182872 | C18H18F3N3O5S | 445.413 | 10 / 2 | N/A | N/A |
| 4797 | 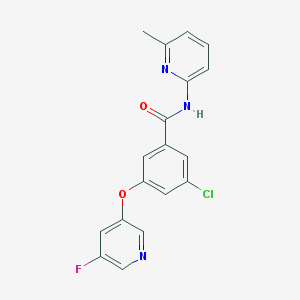 CHEMBL2440638 CHEMBL2440638 | C18H13ClFN3O2 | 357.769 | 5 / 1 | 3.6 | Yes |
| 4945 | 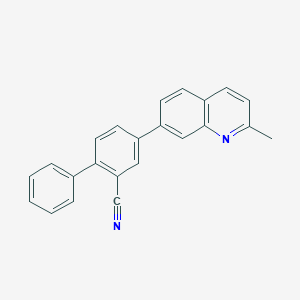 CHEMBL1099113 CHEMBL1099113 | C23H16N2 | 320.395 | 2 / 0 | 5.6 | No |
| 5212 | 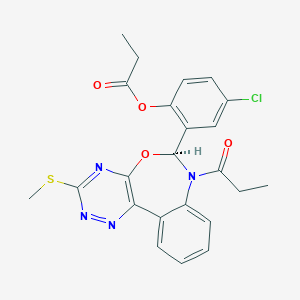 CHEMBL1258628 CHEMBL1258628 | C23H21ClN4O4S | 484.955 | 8 / 0 | 4.1 | Yes |
| 441912 | 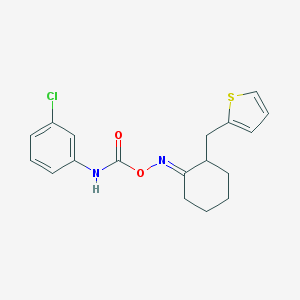 CHEMBL3401568 CHEMBL3401568 | C18H19ClN2O2S | 362.872 | 4 / 1 | 5.0 | Yes |
| 5369 | 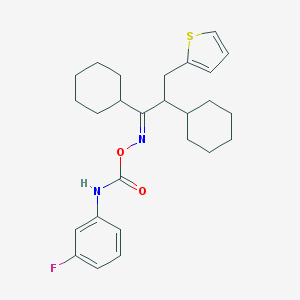 CHEMBL1209477 CHEMBL1209477 | C26H33FN2O2S | 456.62 | 5 / 1 | 8.3 | No |
| 5442 | 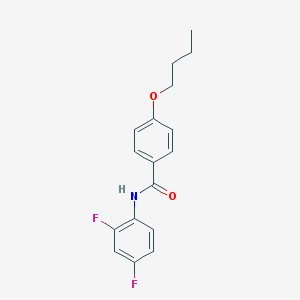 VU 0357121 VU 0357121 | C17H17F2NO2 | 305.325 | 4 / 1 | 4.2 | Yes |
| 459268 | 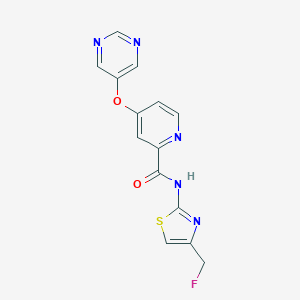 SCHEMBL6929110 SCHEMBL6929110 | C14H10FN5O2S | 331.325 | 8 / 1 | 1.4 | Yes |
| 5493 | 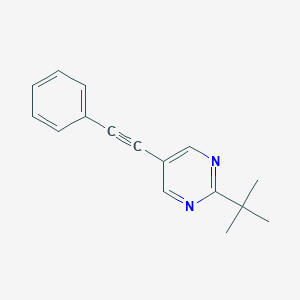 CHEMBL496875 CHEMBL496875 | C16H16N2 | 236.318 | 2 / 0 | 3.9 | Yes |
| 459274 | 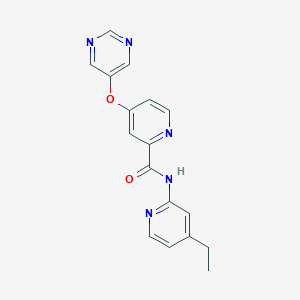 SCHEMBL6928316 SCHEMBL6928316 | C17H15N5O2 | 321.34 | 6 / 1 | 1.9 | Yes |
| 463578 | 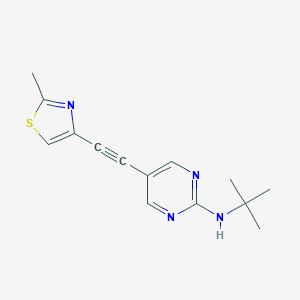 SCHEMBL1244610 SCHEMBL1244610 | C14H16N4S | 272.37 | 5 / 1 | 3.0 | Yes |
| 459276 | 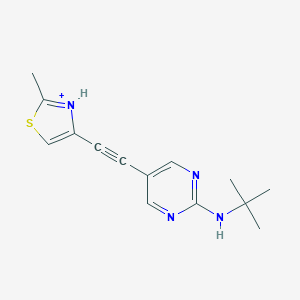 US8609852, 117 US8609852, 117 | C14H17N4S+ | 273.378 | 4 / 2 | 3.2 | Yes |
| 5844 | 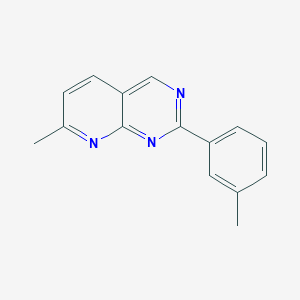 CHEMBL238552 CHEMBL238552 | C15H13N3 | 235.29 | 3 / 0 | 3.2 | Yes |
| 6346 | 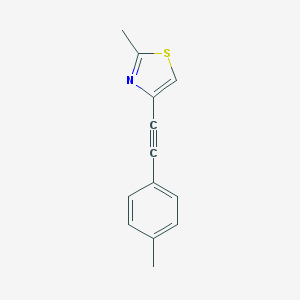 CHEMBL185813 CHEMBL185813 | C13H11NS | 213.298 | 2 / 0 | 3.8 | Yes |
| 6711 | 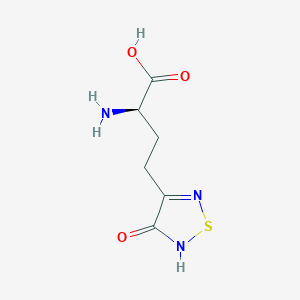 CHEMBL334842 CHEMBL334842 | C6H9N3O3S | 203.216 | 6 / 3 | -3.4 | Yes |
| 6718 | 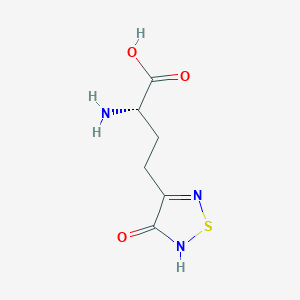 CHEMBL131922 CHEMBL131922 | C6H9N3O3S | 203.216 | 6 / 3 | -3.4 | Yes |
| 6750 | 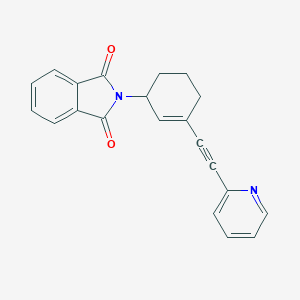 CHEMBL2113099 CHEMBL2113099 | C21H16N2O2 | 328.371 | 3 / 0 | 2.9 | Yes |
| 6830 | 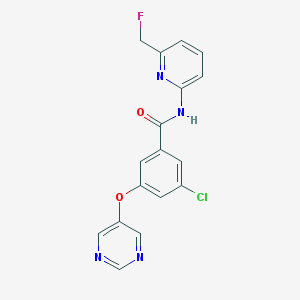 SCHEMBL6918118 SCHEMBL6918118 | C17H12ClFN4O2 | 358.757 | 6 / 1 | 2.6 | Yes |
| 463752 | 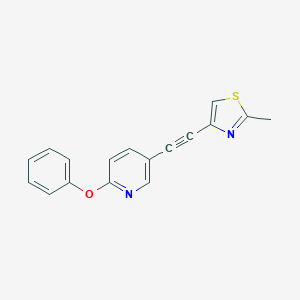 CHEMBL3643058 CHEMBL3643058 | C17H12N2OS | 292.356 | 4 / 0 | 4.2 | Yes |
| 459282 | 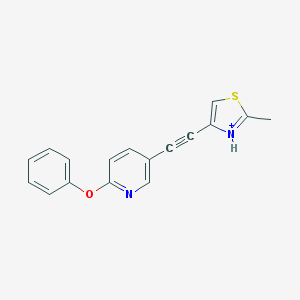 BDBM109171 BDBM109171 | C17H13N2OS+ | 293.364 | 3 / 1 | 4.4 | Yes |
| 7332 | 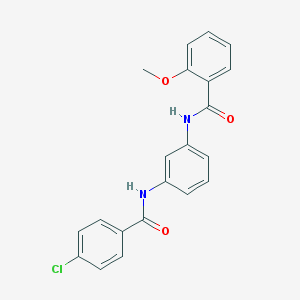 CHEMBL1082076 CHEMBL1082076 | C21H17ClN2O3 | 380.828 | 3 / 2 | 5.0 | Yes |
| 7428 | 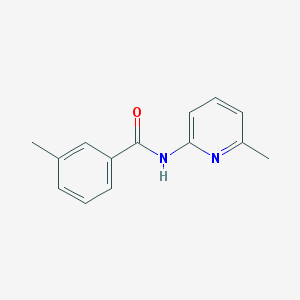 3-methyl-N-(6-methylpyridin-2-yl)benzamide 3-methyl-N-(6-methylpyridin-2-yl)benzamide | C14H14N2O | 226.279 | 2 / 1 | 2.8 | Yes |
| 557500 | 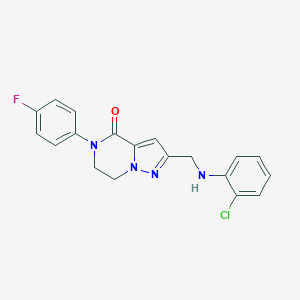 SCHEMBL17334341 SCHEMBL17334341 | C19H16ClFN4O | 370.812 | 4 / 1 | 3.5 | Yes |
| 7993 | 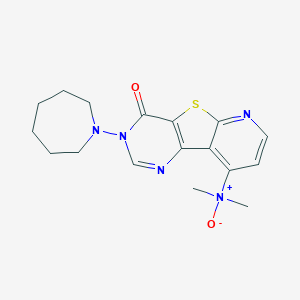 CHEMBL386408 CHEMBL386408 | C17H21N5O2S | 359.448 | 6 / 0 | 2.3 | Yes |
| 8264 | 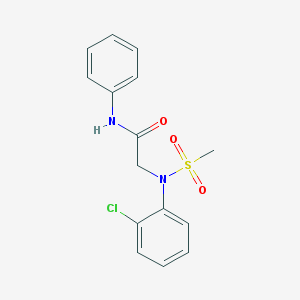 CHEMBL2208408 CHEMBL2208408 | C15H15ClN2O3S | 338.806 | 4 / 1 | 2.6 | Yes |
| 8347 | 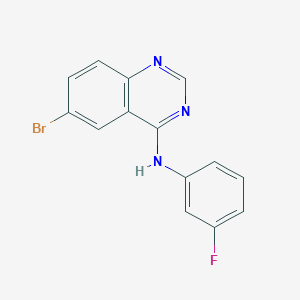 CHEMBL1079375 CHEMBL1079375 | C14H9BrFN3 | 318.149 | 4 / 1 | 4.2 | Yes |
| 8566 | 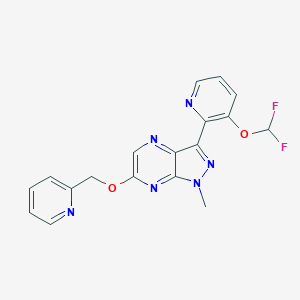 CHEMBL3122214 CHEMBL3122214 | C18H14F2N6O2 | 384.347 | 9 / 0 | 2.2 | Yes |
| 8768 | 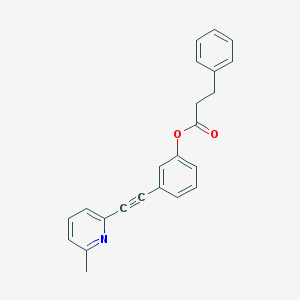 CHEMBL185333 CHEMBL185333 | C23H19NO2 | 341.41 | 3 / 0 | 5.0 | Yes |
| 9357 | 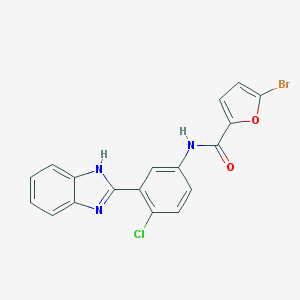 CHEMBL1081892 CHEMBL1081892 | C18H11BrClN3O2 | 416.659 | 3 / 2 | 5.0 | Yes |
| 464030 | 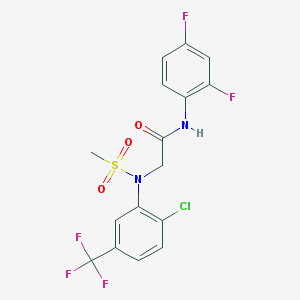 478067-38-8 478067-38-8 | C16H12ClF5N2O3S | 442.785 | 9 / 1 | 3.7 | Yes |
| 9629 | 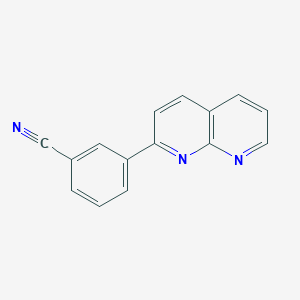 CHEMBL235397 CHEMBL235397 | C15H9N3 | 231.258 | 3 / 0 | 2.8 | Yes |
| 9651 | 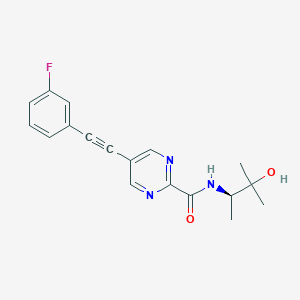 CHEMBL2431153 CHEMBL2431153 | C18H18FN3O2 | 327.359 | 5 / 2 | 2.3 | Yes |
| 9728 | 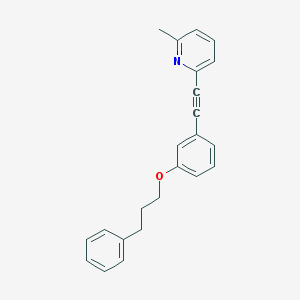 CHEMBL182019 CHEMBL182019 | C23H21NO | 327.427 | 2 / 0 | 5.6 | No |
| 459307 | 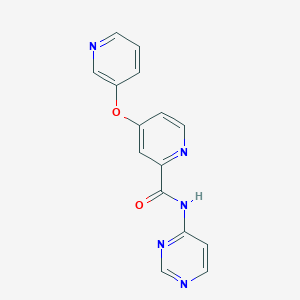 SCHEMBL6929148 SCHEMBL6929148 | C15H11N5O2 | 293.286 | 6 / 1 | 1.1 | Yes |
| 10329 |  CHEMBL2431204 CHEMBL2431204 | C18H24N2O2 | 300.402 | 3 / 2 | 3.2 | Yes |
| 10349 | 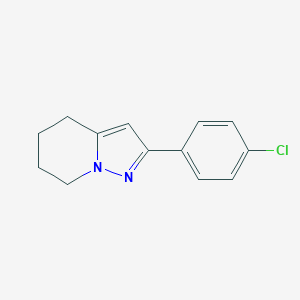 CHEMBL2408177 CHEMBL2408177 | C13H13ClN2 | 232.711 | 1 / 0 | 3.2 | Yes |
| 10398 | 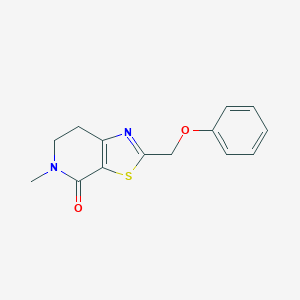 CHEMBL2426616 CHEMBL2426616 | C14H14N2O2S | 274.338 | 4 / 0 | 2.3 | Yes |
| 10535 | 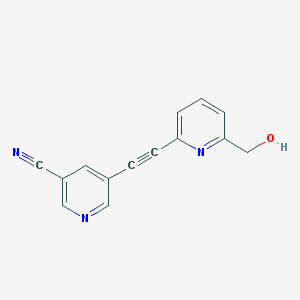 CHEMBL2089188 CHEMBL2089188 | C14H9N3O | 235.246 | 4 / 1 | 0.7 | Yes |
| 10658 | 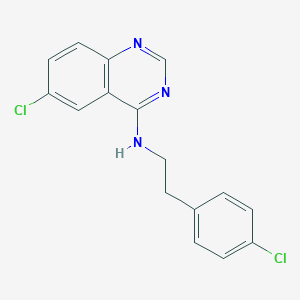 CHEMBL149800 CHEMBL149800 | C16H13Cl2N3 | 318.201 | 3 / 1 | 5.1 | No |
| 10752 | 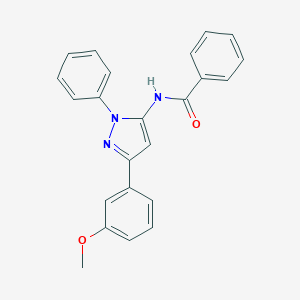 CHEMBL445149 CHEMBL445149 | C23H19N3O2 | 369.424 | 3 / 1 | 4.7 | Yes |
| 10879 | 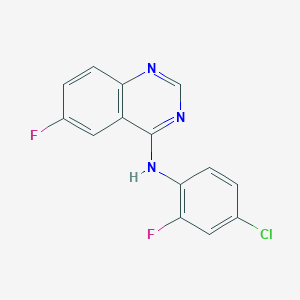 CHEMBL1079404 CHEMBL1079404 | C14H8ClF2N3 | 291.686 | 5 / 1 | 4.2 | Yes |
| 10967 | 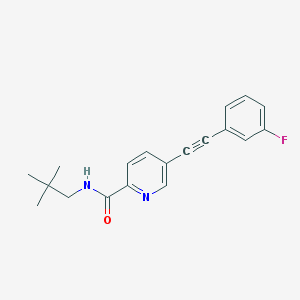 CHEMBL2431164 CHEMBL2431164 | C19H19FN2O | 310.372 | 3 / 1 | 4.3 | Yes |
| 11175 | 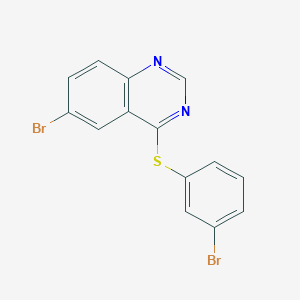 CHEMBL1079934 CHEMBL1079934 | C14H8Br2N2S | 396.1 | 3 / 0 | 5.3 | No |
| 521770 | 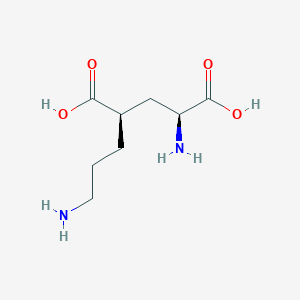 CHEMBL2312685 CHEMBL2312685 | C8H16N2O4 | 204.226 | 6 / 4 | -6.8 | Yes |
| 442155 |  CHEMBL3401198 CHEMBL3401198 | C18H15FN4O2 | 338.342 | 5 / 0 | 2.4 | Yes |
| 442158 | 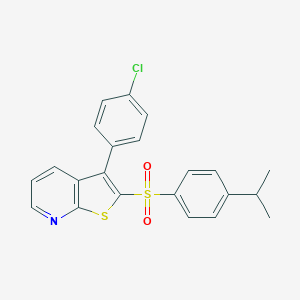 CHEMBL3401583 CHEMBL3401583 | C22H18ClNO2S2 | 427.961 | 4 / 0 | 6.7 | No |
| 12268 | 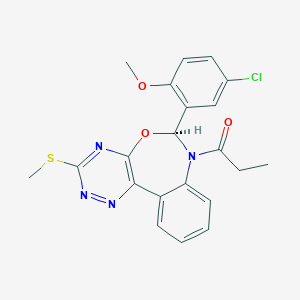 CHEMBL1258403 CHEMBL1258403 | C21H19ClN4O3S | 442.918 | 7 / 0 | 3.8 | Yes |
| 459329 | 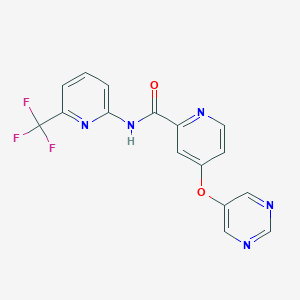 SCHEMBL6927274 SCHEMBL6927274 | C16H10F3N5O2 | 361.284 | 9 / 1 | 2.0 | Yes |
| 464441 | 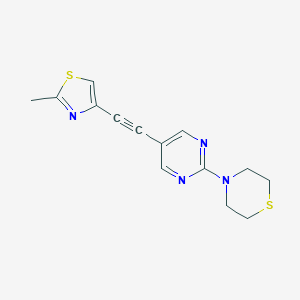 CHEMBL3645567 CHEMBL3645567 | C14H14N4S2 | 302.414 | 6 / 0 | 2.6 | Yes |
| 459338 | 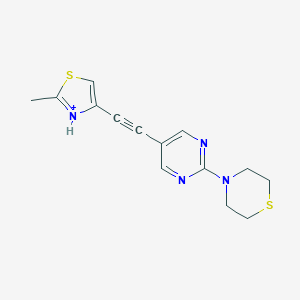 BDBM109191 BDBM109191 | C14H15N4S2+ | 303.422 | 5 / 1 | 2.7 | Yes |
| 13081 | 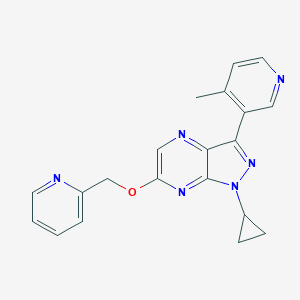 CHEMBL3122219 CHEMBL3122219 | C20H18N6O | 358.405 | 6 / 0 | 2.0 | Yes |
| 13316 | 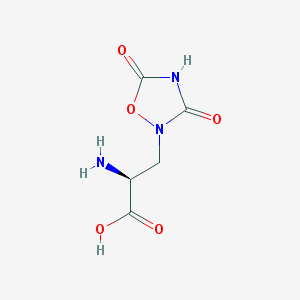 QUISQUALIC ACID QUISQUALIC ACID | C5H7N3O5 | 189.127 | 6 / 3 | -3.9 | Yes |
| 555531 | 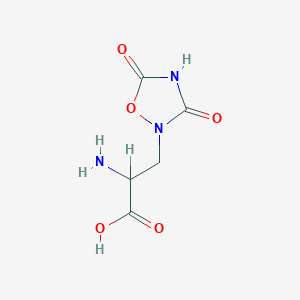 2-amino-3-(3,5-dioxo-1,2,4-oxadiazolidin-2-yl)propanoic acid 2-amino-3-(3,5-dioxo-1,2,4-oxadiazolidin-2-yl)propanoic acid | C5H7N3O5 | 189.127 | 6 / 3 | -3.9 | Yes |
| 464487 | 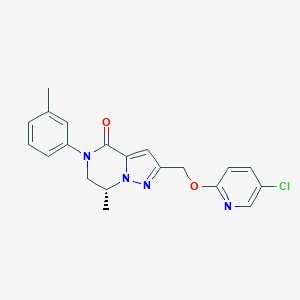 CHEMBL3617636 CHEMBL3617636 | C20H19ClN4O2 | 382.848 | 4 / 0 | 3.5 | Yes |
| 13880 |  CHEMBL363062 CHEMBL363062 | C22H16ClN3O | 373.84 | 2 / 1 | 5.3 | No |
| 13902 | 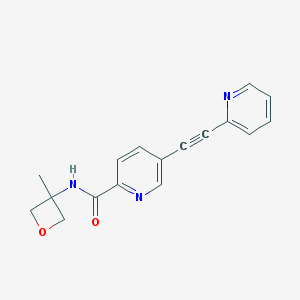 CHEMBL2431183 CHEMBL2431183 | C17H15N3O2 | 293.326 | 4 / 1 | 1.3 | Yes |
| 14016 |  CHEMBL561189 CHEMBL561189 | C19H14ClNO | 307.777 | 2 / 0 | 5.3 | No |
| 464658 |  CHEMBL3617514 CHEMBL3617514 | C19H16F2N4O2 | 370.36 | 6 / 0 | 2.7 | Yes |
| 14806 | 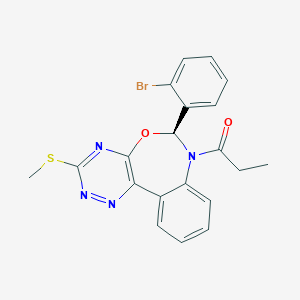 CHEMBL1257238 CHEMBL1257238 | C20H17BrN4O2S | 457.346 | 6 / 0 | 3.9 | Yes |
| 14891 |  CHEMBL362217 CHEMBL362217 | C16H11N3S | 277.345 | 4 / 0 | 3.0 | Yes |
| 14949 | 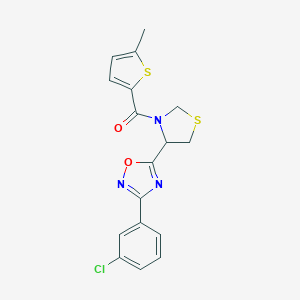 CHEMBL1083340 CHEMBL1083340 | C17H14ClN3O2S2 | 391.888 | 6 / 0 | 4.4 | Yes |
| 442241 | 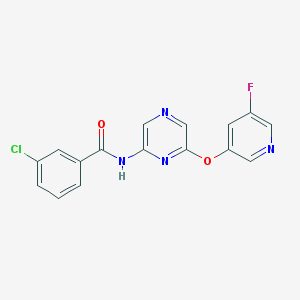 CHEMBL3314817 CHEMBL3314817 | C16H10ClFN4O2 | 344.73 | 6 / 1 | 2.5 | Yes |
| 15215 | 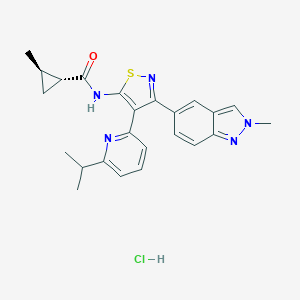 CHEMBL2334981 CHEMBL2334981 | C24H26ClN5OS | 468.016 | 5 / 2 | N/A | N/A |
| 15295 | 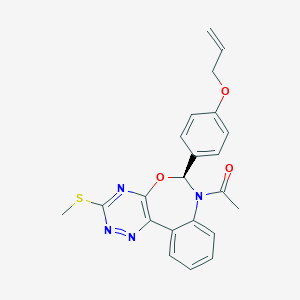 CHEMBL1257467 CHEMBL1257467 | C22H20N4O3S | 420.487 | 7 / 0 | 3.4 | Yes |
| 15347 | 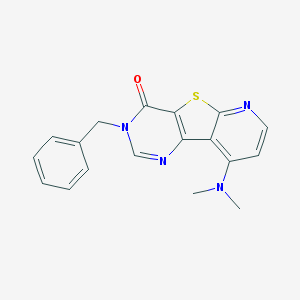 CHEMBL388087 CHEMBL388087 | C18H16N4OS | 336.413 | 5 / 0 | 3.1 | Yes |
| 15362 | 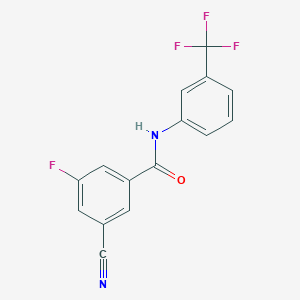 CHEMBL1209547 CHEMBL1209547 | C15H8F4N2O | 308.236 | 6 / 1 | 4.0 | Yes |
| 553327 |  methoxymethyl-MTEP methoxymethyl-MTEP | C13H12N2OS | 244.312 | 4 / 0 | 2.0 | Yes |
| 16000 | 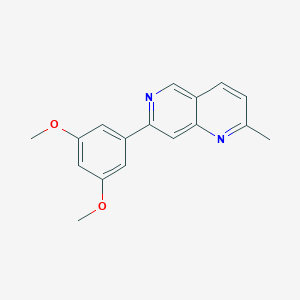 CHEMBL238344 CHEMBL238344 | C17H16N2O2 | 280.327 | 4 / 0 | 3.1 | Yes |
| 16025 | 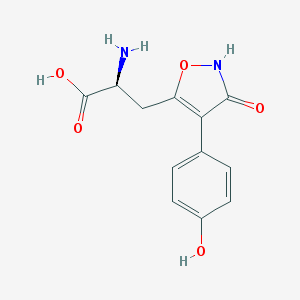 CHEMBL367027 CHEMBL367027 | C12H12N2O5 | 264.237 | 6 / 4 | -2.3 | Yes |
| 16142 | 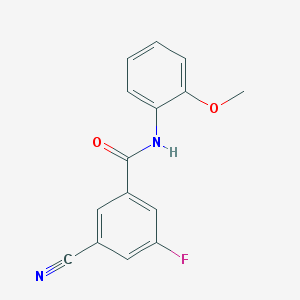 CHEMBL1209469 CHEMBL1209469 | C15H11FN2O2 | 270.263 | 4 / 1 | 2.5 | Yes |
| 442295 |  CHEMBL3314838 CHEMBL3314838 | C13H8FN5O2S | 317.298 | 8 / 1 | 1.2 | Yes |
| 16445 | 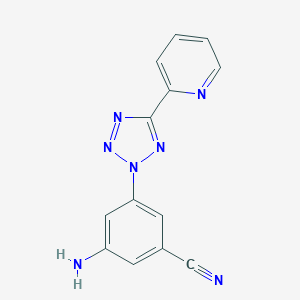 CHEMBL372809 CHEMBL372809 | C13H9N7 | 263.264 | 6 / 1 | 1.4 | Yes |
| 16503 | 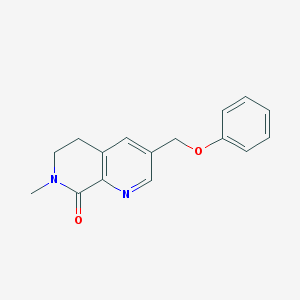 CHEMBL3298467 CHEMBL3298467 | C16H16N2O2 | 268.316 | 3 / 0 | 2.1 | Yes |
| 16545 | 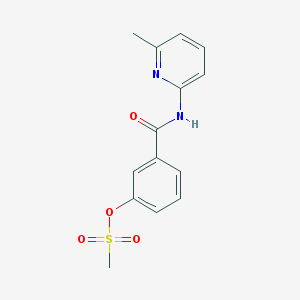 CHEMBL210512 CHEMBL210512 | C14H14N2O4S | 306.336 | 5 / 1 | 2.0 | Yes |
| 464946 | 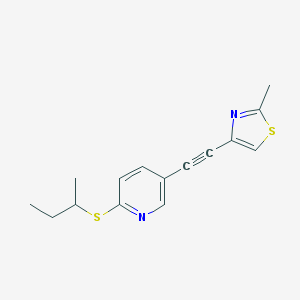 SCHEMBL8234690 SCHEMBL8234690 | C15H16N2S2 | 288.427 | 4 / 0 | 4.5 | Yes |
| 459360 | 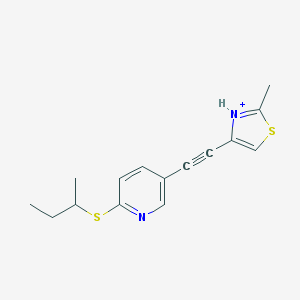 BDBM109174 BDBM109174 | C15H17N2S2+ | 289.435 | 3 / 1 | 4.7 | Yes |
| 17122 | 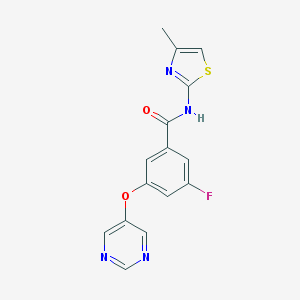 CHEMBL2440659 CHEMBL2440659 | C15H11FN4O2S | 330.337 | 7 / 1 | 2.4 | Yes |
| 17197 |  CHEMBL1688373 CHEMBL1688373 | C21H24N2O4 | 368.433 | 5 / 0 | 2.5 | Yes |
| 17278 | 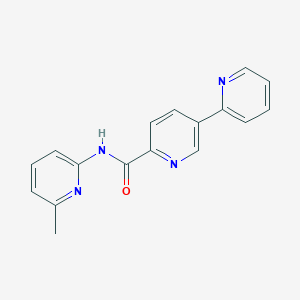 CHEMBL248141 CHEMBL248141 | C17H14N4O | 290.326 | 4 / 1 | 2.3 | Yes |
| 17493 | 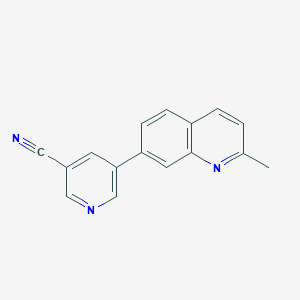 CHEMBL233418 CHEMBL233418 | C16H11N3 | 245.285 | 3 / 0 | 2.9 | Yes |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218