

You can:
| Name | Extracellular calcium-sensing receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | CASR |
| Synonym | hCasR {ECO:0000303|PubMed:27386547} GPRC2A extracellular calcium-sensing receptor Parathyroid cell calcium-sensing receptor 1 {ECO:0000303|PubMed:8698326} PCaR1 {ECO:0000303|PubMed:8698326} [ Show all ] |
| Disease | Osteoporosis Cerebrovascular ischaemia Cardiovascular disease; Kidney disease Secondary hyperparathyroidism Myelodysplastic syndrome [ Show all ] |
| Length | 1078 |
| Amino acid sequence | MAFYSCCWVLLALTWHTSAYGPDQRAQKKGDIILGGLFPIHFGVAAKDQDLKSRPESVECIRYNFRGFRWLQAMIFAIEEINSSPALLPNLTLGYRIFDTCNTVSKALEATLSFVAQNKIDSLNLDEFCNCSEHIPSTIAVVGATGSGVSTAVANLLGLFYIPQVSYASSSRLLSNKNQFKSFLRTIPNDEHQATAMADIIEYFRWNWVGTIAADDDYGRPGIEKFREEAEERDICIDFSELISQYSDEEEIQHVVEVIQNSTAKVIVVFSSGPDLEPLIKEIVRRNITGKIWLASEAWASSSLIAMPQYFHVVGGTIGFALKAGQIPGFREFLKKVHPRKSVHNGFAKEFWEETFNCHLQEGAKGPLPVDTFLRGHEESGDRFSNSSTAFRPLCTGDENISSVETPYIDYTHLRISYNVYLAVYSIAHALQDIYTCLPGRGLFTNGSCADIKKVEAWQVLKHLRHLNFTNNMGEQVTFDECGDLVGNYSIINWHLSPEDGSIVFKEVGYYNVYAKKGERLFINEEKILWSGFSREVPFSNCSRDCLAGTRKGIIEGEPTCCFECVECPDGEYSDETDASACNKCPDDFWSNENHTSCIAKEIEFLSWTEPFGIALTLFAVLGIFLTAFVLGVFIKFRNTPIVKATNRELSYLLLFSLLCCFSSSLFFIGEPQDWTCRLRQPAFGISFVLCISCILVKTNRVLLVFEAKIPTSFHRKWWGLNLQFLLVFLCTFMQIVICVIWLYTAPPSSYRNQELEDEIIFITCHEGSLMALGFLIGYTCLLAAICFFFAFKSRKLPENFNEAKFITFSMLIFFIVWISFIPAYASTYGKFVSAVEVIAILAASFGLLACIFFNKIYIILFKPSRNTIEEVRCSTAAHAFKVAARATLRRSNVSRKRSSSLGGSTGSTPSSSISSKSNSEDPFPQPERQKQQQPLALTQQEQQQQPLTLPQQQRSQQQPRCKQKVIFGSGTVTFSLSFDEPQKNAMAHRNSTHQNSLEAQKSSDTLTRHEPLLPLQCGETDLDLTVQETGLQGPVGGDQRPEVEDPEELSPALVVSSSQSFVISGGGSTVTENVVNS |
| UniProt | P41180 |
| Protein Data Bank | 5k5t, 5k5s, 5fbk, 5fbh |
| GPCR-HGmod model | N/A |
| 3D structure model | This structure is from PDB ID 5k5t. |
| BioLiP | BL0353424,BL0353430, BL0353434, BL0353425,BL0353426,BL0353427,, BL0349612,BL0349615, BL0349611,BL0349614, BL0349610,BL0349613, BL0349606,BL0349609, BL0349605,BL0349608, BL0349604,BL0349607 |
| Therapeutic Target Database | T92076 |
| ChEMBL | CHEMBL1878 |
| IUPHAR | 54 |
| DrugBank | BE0000509 |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 521438 | 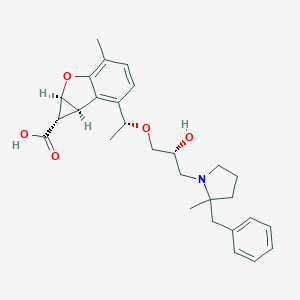 CHEMBL3828329 CHEMBL3828329 | C28H35NO5 | 465.59 | 6 / 2 | 1.3 | Yes |
| 108 | 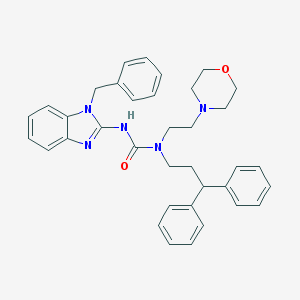 CHEMBL2377738 CHEMBL2377738 | C36H39N5O2 | 573.741 | 4 / 1 | 6.0 | No |
| 151 | 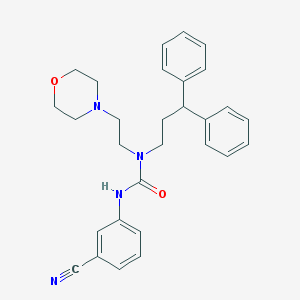 CHEMBL2349596 CHEMBL2349596 | C29H32N4O2 | 468.601 | 4 / 1 | 4.3 | Yes |
| 557328 | 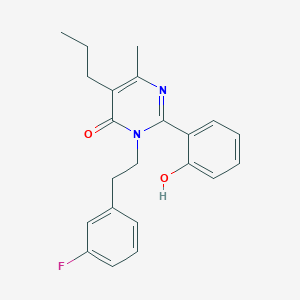 CHEMBL195566 CHEMBL195566 | C22H23FN2O2 | 366.436 | 4 / 1 | 4.5 | Yes |
| 557334 |  CHEMBL1689044 CHEMBL1689044 | C33H38N4O | 506.694 | 3 / 2 | 7.2 | No |
| 1943 |  CHEMBL458516 CHEMBL458516 | C28H27N5O6 | 529.553 | 10 / 2 | 4.0 | No |
| 3220 |  CHEMBL2346767 CHEMBL2346767 | C23H24N2O2 | 360.457 | 2 / 2 | 4.7 | Yes |
| 4013 |  CHEMBL599039 CHEMBL599039 | C26H35N3O | 405.586 | 2 / 0 | 6.7 | No |
| 557405 |  CHEMBL1688085 CHEMBL1688085 | C26H31FN4O | 434.559 | 4 / 2 | 5.3 | No |
| 5820 | 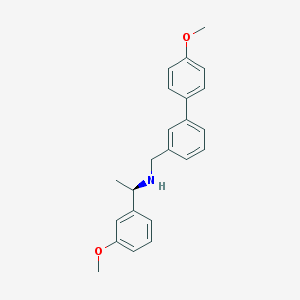 CHEMBL1224263 CHEMBL1224263 | C23H25NO2 | 347.458 | 3 / 1 | 4.8 | Yes |
| 536091 | 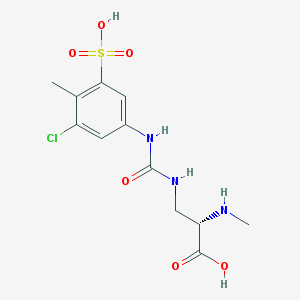 (2S)-3-{[(3-chloro-4-methyl-5-sulfophenyl)carbamoyl]amino}-2-(methylamino)propanoic acid (2S)-3-{[(3-chloro-4-methyl-5-sulfophenyl)carbamoyl]amino}-2-(methylamino)propanoic acid | C12H16ClN3O6S | 365.785 | 7 / 5 | -2.4 | Yes |
| 557477 | 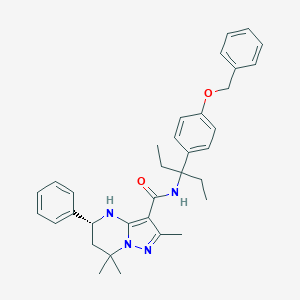 CHEMBL1689055 CHEMBL1689055 | C34H40N4O2 | 536.72 | 4 / 2 | 7.0 | No |
| 521660 | 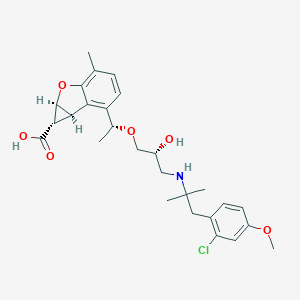 CHEMBL3828187 CHEMBL3828187 | C27H34ClNO6 | 504.02 | 7 / 3 | 1.4 | No |
| 7727 |  CHEMBL1224260 CHEMBL1224260 | C22H22FNO | 335.422 | 3 / 1 | 4.9 | Yes |
| 9023 | 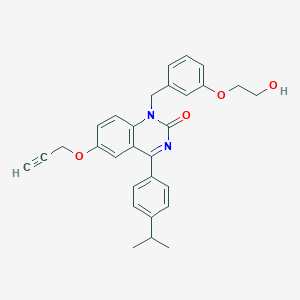 CHEMBL597396 CHEMBL597396 | C29H28N2O4 | 468.553 | 4 / 1 | 4.9 | Yes |
| 547994 |  etelcalcetide etelcalcetide | C38H73N21O10S2 | 1048.26 | 17 / 18 | -10.7 | No |
| 557572 | 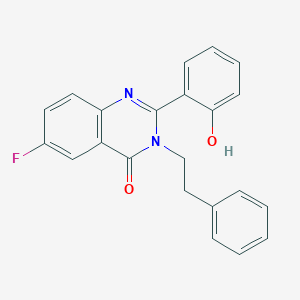 CHEMBL183287 CHEMBL183287 | C22H17FN2O2 | 360.388 | 4 / 1 | 4.2 | Yes |
| 10902 | 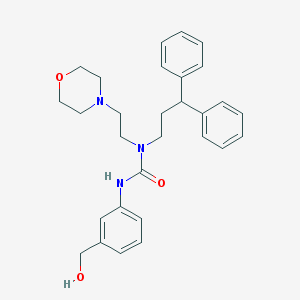 CHEMBL2349588 CHEMBL2349588 | C29H35N3O3 | 473.617 | 4 / 2 | 3.7 | Yes |
| 12107 |  CHEMBL199548 CHEMBL199548 | C28H33N3O | 427.592 | 4 / 2 | 4.8 | Yes |
| 12451 | 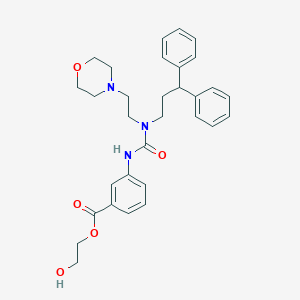 CHEMBL2349584 CHEMBL2349584 | C31H37N3O5 | 531.653 | 6 / 2 | 3.7 | No |
| 12866 | 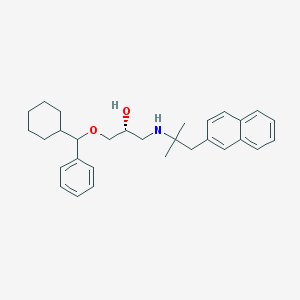 CHEMBL1084201 CHEMBL1084201 | C30H39NO2 | 445.647 | 3 / 2 | 6.8 | No |
| 557653 | 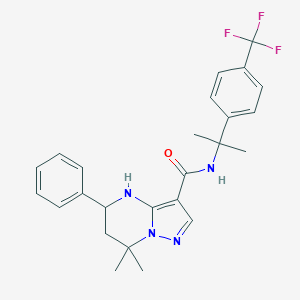 CHEMBL1689809 CHEMBL1689809 | C25H27F3N4O | 456.513 | 6 / 2 | 5.0 | Yes |
| 521870 | 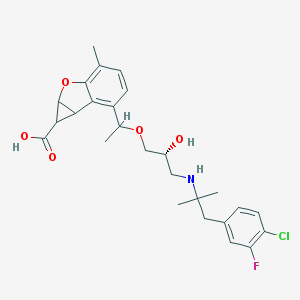 CHEMBL3828182 CHEMBL3828182 | C26H31ClFNO5 | 491.984 | 7 / 3 | 1.5 | Yes |
| 521871 | 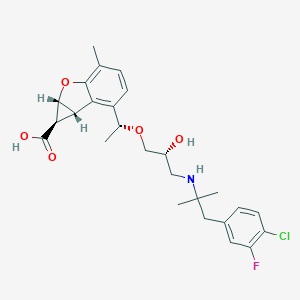 CHEMBL3827356 CHEMBL3827356 | C26H31ClFNO5 | 491.984 | 7 / 3 | 1.5 | Yes |
| 521872 | 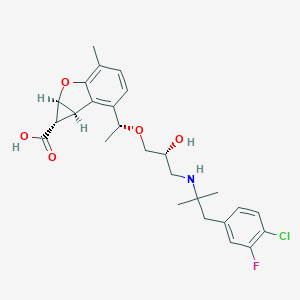 CHEMBL3827154 CHEMBL3827154 | C26H31ClFNO5 | 491.984 | 7 / 3 | 1.5 | Yes |
| 14261 | 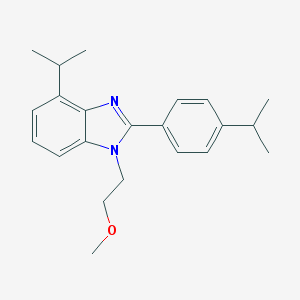 CHEMBL1223222 CHEMBL1223222 | C22H28N2O | 336.479 | 2 / 0 | 5.1 | No |
| 521885 | 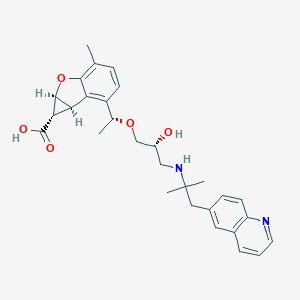 CHEMBL3827410 CHEMBL3827410 | C29H34N2O5 | 490.6 | 7 / 3 | 1.0 | Yes |
| 14887 | 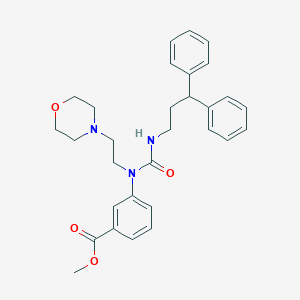 CHEMBL2346781 CHEMBL2346781 | C30H35N3O4 | 501.627 | 5 / 1 | 4.4 | No |
| 15974 |  CHEMBL1224190 CHEMBL1224190 | C16H19NO | 241.334 | 2 / 1 | 3.2 | Yes |
| 557750 | 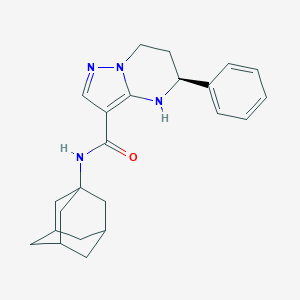 CHEMBL1290216 CHEMBL1290216 | C23H28N4O | 376.504 | 3 / 2 | 4.1 | Yes |
| 17022 | 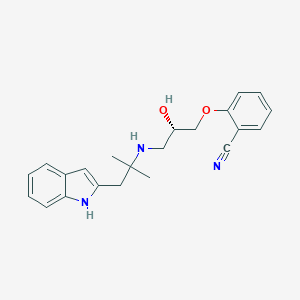 CHEMBL371936 CHEMBL371936 | C22H25N3O2 | 363.461 | 4 / 3 | 3.3 | Yes |
| 18917 | 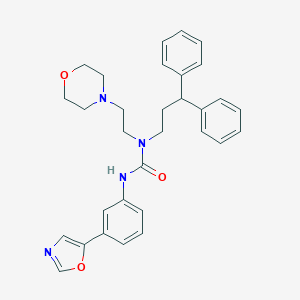 CHEMBL2349603 CHEMBL2349603 | C31H34N4O3 | 510.638 | 5 / 1 | 4.6 | No |
| 557857 | 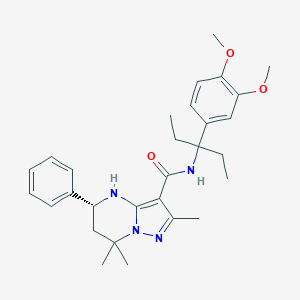 CHEMBL1689054 CHEMBL1689054 | C29H38N4O3 | 490.648 | 5 / 2 | 5.5 | No |
| 20228 |  CHEMBL1223299 CHEMBL1223299 | C19H22N2O | 294.398 | 2 / 0 | 4.0 | Yes |
| 20422 | 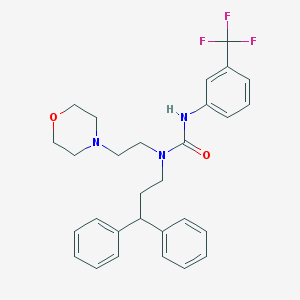 CHEMBL2349597 CHEMBL2349597 | C29H32F3N3O2 | 511.589 | 6 / 1 | 5.4 | No |
| 20805 | 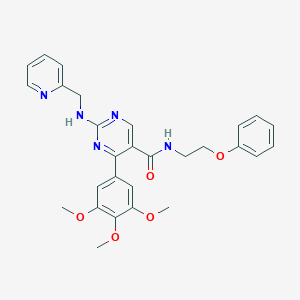 CHEMBL490492 CHEMBL490492 | C28H29N5O5 | 515.57 | 9 / 2 | 3.5 | No |
| 21055 | 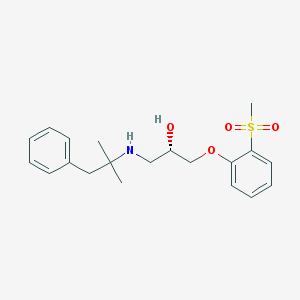 CHEMBL200248 CHEMBL200248 | C20H27NO4S | 377.499 | 5 / 2 | 2.7 | Yes |
| 536511 | 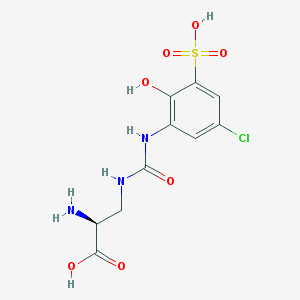 CHEMBL3980928 CHEMBL3980928 | C10H12ClN3O7S | 353.73 | 8 / 6 | -3.1 | No |
| 21509 | 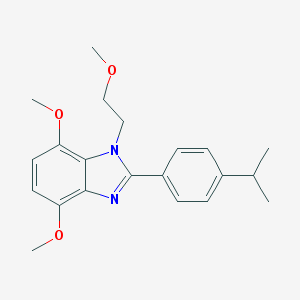 CHEMBL1223370 CHEMBL1223370 | C21H26N2O3 | 354.45 | 4 / 0 | 4.0 | Yes |
| 22226 | 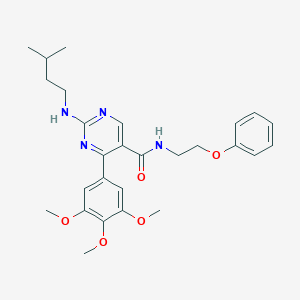 CHEMBL454375 CHEMBL454375 | C27H34N4O5 | 494.592 | 8 / 2 | 4.7 | Yes |
| 522342 | 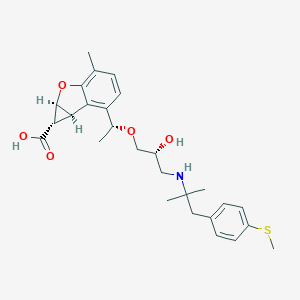 CHEMBL3827209 CHEMBL3827209 | C27H35NO5S | 485.639 | 7 / 3 | 1.3 | Yes |
| 25934 | 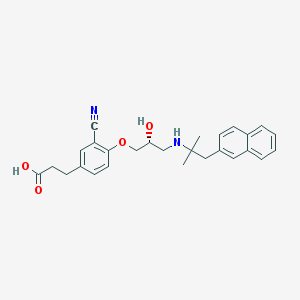 CHEMBL570593 CHEMBL570593 | C27H30N2O4 | 446.547 | 6 / 3 | 1.9 | Yes |
| 536692 | 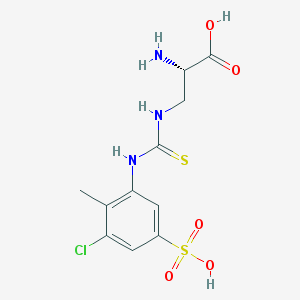 CHEMBL3895461 CHEMBL3895461 | C11H14ClN3O5S2 | 367.819 | 7 / 5 | -2.3 | Yes |
| 28186 | 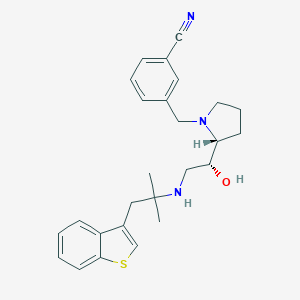 CHEMBL200041 CHEMBL200041 | C26H31N3OS | 433.614 | 5 / 2 | 4.6 | Yes |
| 28923 | 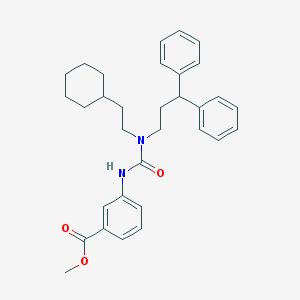 CHEMBL2346785 CHEMBL2346785 | C32H38N2O3 | 498.667 | 3 / 1 | 7.8 | No |
| 29318 | 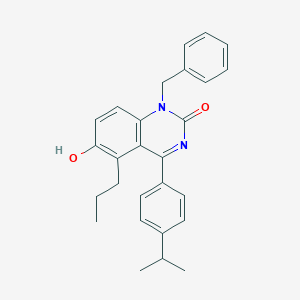 CHEMBL598016 CHEMBL598016 | C27H28N2O2 | 412.533 | 2 / 1 | 6.5 | No |
| 31780 | 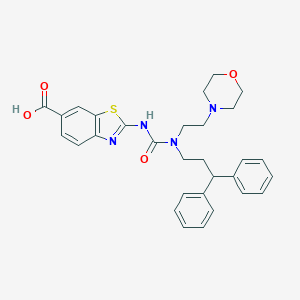 CHEMBL2375383 CHEMBL2375383 | C30H32N4O4S | 544.67 | 7 / 2 | 2.6 | No |
| 32736 |  CHEMBL180549 CHEMBL180549 | C22H17FN2O | 344.389 | 3 / 0 | 4.6 | Yes |
| 33454 | 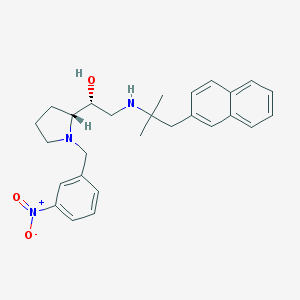 CHEMBL372820 CHEMBL372820 | C27H33N3O3 | 447.579 | 5 / 2 | 4.9 | Yes |
| 558353 | 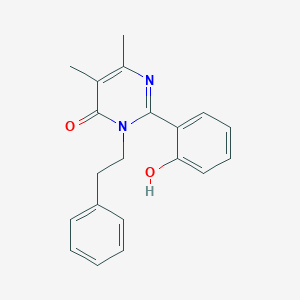 CHEMBL371828 CHEMBL371828 | C20H20N2O2 | 320.392 | 3 / 1 | 3.5 | Yes |
| 35316 | 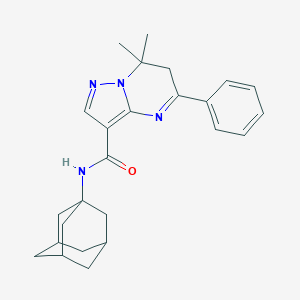 BDBM50331965 BDBM50331965 | C25H30N4O | 402.542 | 3 / 1 | 4.2 | Yes |
| 36009 | 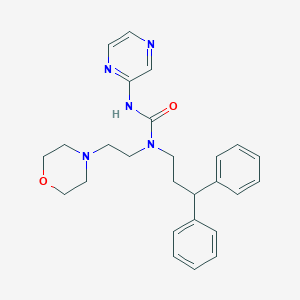 CHEMBL2377728 CHEMBL2377728 | C26H31N5O2 | 445.567 | 5 / 1 | 2.7 | Yes |
| 37636 | 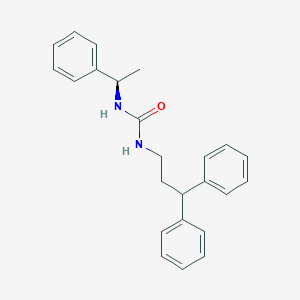 CHEMBL2346666 CHEMBL2346666 | C24H26N2O | 358.485 | 1 / 2 | 5.0 | Yes |
| 38821 | 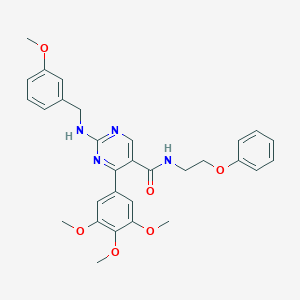 CHEMBL493228 CHEMBL493228 | C30H32N4O6 | 544.608 | 9 / 2 | 4.5 | No |
| 38878 | 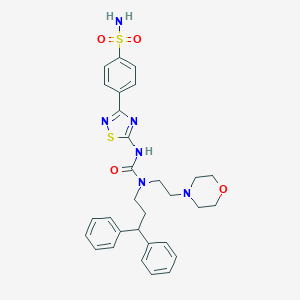 CHEMBL3093424 CHEMBL3093424 | C30H34N6O4S2 | 606.76 | 9 / 2 | 3.9 | No |
| 536992 | 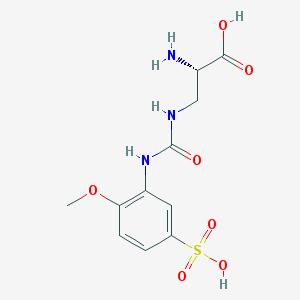 CHEMBL3964061 CHEMBL3964061 | C11H15N3O7S | 333.315 | 8 / 5 | -3.9 | Yes |
| 39634 | 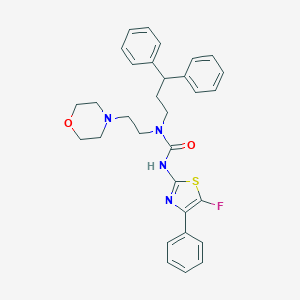 CHEMBL3093418 CHEMBL3093418 | C31H33FN4O2S | 544.689 | 6 / 1 | 6.0 | No |
| 40742 | 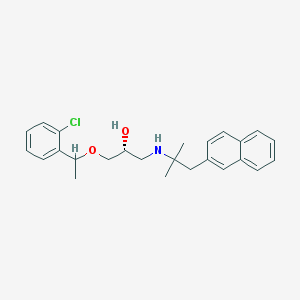 CHEMBL1083307 CHEMBL1083307 | C25H30ClNO2 | 411.97 | 3 / 2 | 5.4 | No |
| 41306 | 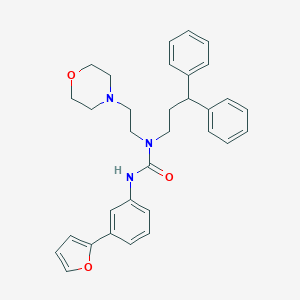 CHEMBL2349602 CHEMBL2349602 | C32H35N3O3 | 509.65 | 4 / 1 | 5.3 | No |
| 42147 | 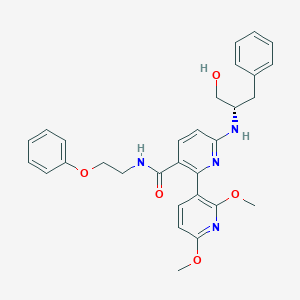 802916-41-2 802916-41-2 | C30H32N4O5 | 528.609 | 8 / 3 | 4.7 | No |
| 42712 | 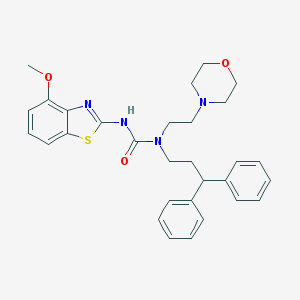 CHEMBL2377741 CHEMBL2377741 | C30H34N4O3S | 530.687 | 6 / 1 | 5.3 | No |
| 43164 | 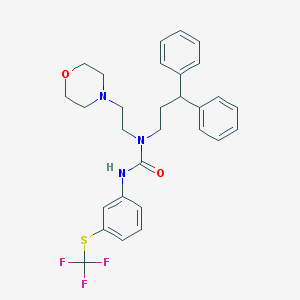 CHEMBL2349591 CHEMBL2349591 | C29H32F3N3O2S | 543.649 | 7 / 1 | 6.3 | No |
| 43226 |  CHEMBL1082711 CHEMBL1082711 | C26H33NO2 | 391.555 | 3 / 2 | 5.3 | No |
| 44750 | 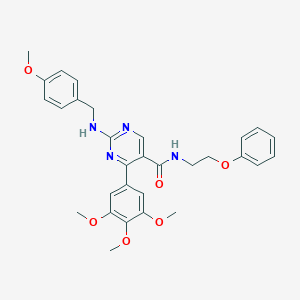 CHEMBL523682 CHEMBL523682 | C30H32N4O6 | 544.608 | 9 / 2 | 4.5 | No |
| 45156 | 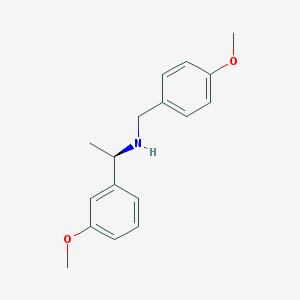 CHEMBL1224193 CHEMBL1224193 | C17H21NO2 | 271.36 | 3 / 1 | 3.1 | Yes |
| 46657 |  CHEMBL599665 CHEMBL599665 | C21H22N2O2 | 334.419 | 2 / 0 | 4.5 | Yes |
| 47317 |  CHEMBL1256301 CHEMBL1256301 | C15H12ClNOS | 289.777 | 3 / 1 | 4.0 | Yes |
| 558777 | 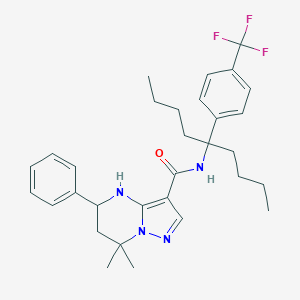 CHEMBL1689812 CHEMBL1689812 | C31H39F3N4O | 540.675 | 6 / 2 | 7.8 | No |
| 51140 | 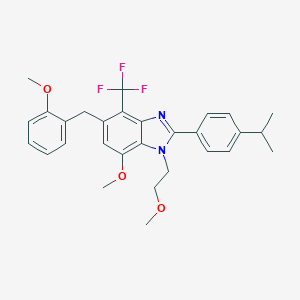 CHEMBL1222489 CHEMBL1222489 | C29H31F3N2O3 | 512.573 | 7 / 0 | 6.8 | No |
| 51591 | 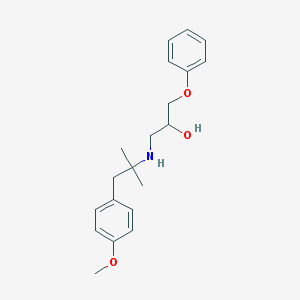 CHEMBL488337 CHEMBL488337 | C20H27NO3 | 329.44 | 4 / 2 | 3.4 | Yes |
| 558875 | 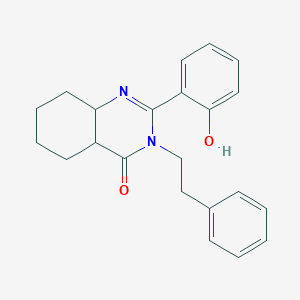 CHEMBL369998 CHEMBL369998 | C22H24N2O2 | 348.446 | 3 / 1 | 3.8 | Yes |
| 558881 |  CHEMBL1289679 CHEMBL1289679 | C24H26ClF3N4O | 478.944 | 6 / 2 | 5.9 | No |
| 558904 | 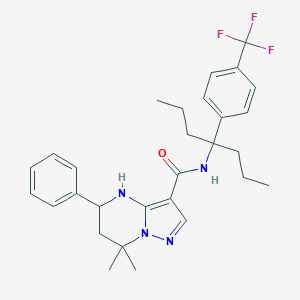 CHEMBL1689811 CHEMBL1689811 | C29H35F3N4O | 512.621 | 6 / 2 | 6.7 | No |
| 53163 | 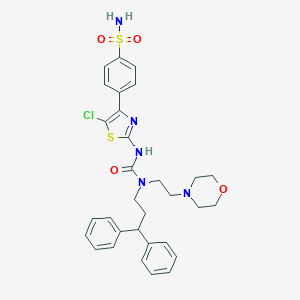 CHEMBL3093422 CHEMBL3093422 | C31H34ClN5O4S2 | 640.214 | 8 / 2 | 5.1 | No |
| 53226 | 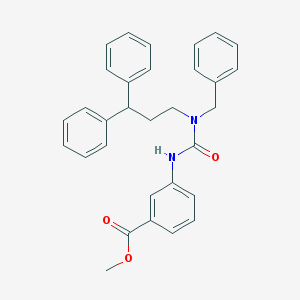 CHEMBL2346783 CHEMBL2346783 | C31H30N2O3 | 478.592 | 3 / 1 | 6.2 | No |
| 54245 | 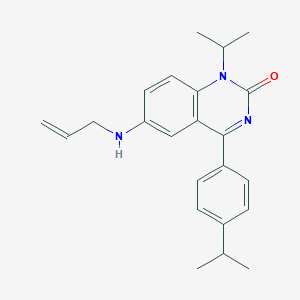 CHEMBL599040 CHEMBL599040 | C23H27N3O | 361.489 | 2 / 1 | 5.4 | No |
| 54384 | 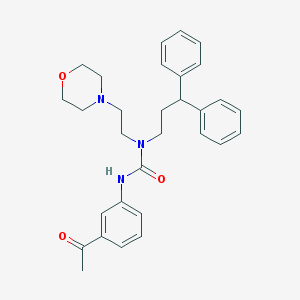 CHEMBL2349587 CHEMBL2349587 | C30H35N3O3 | 485.628 | 4 / 1 | 4.2 | Yes |
| 55936 | 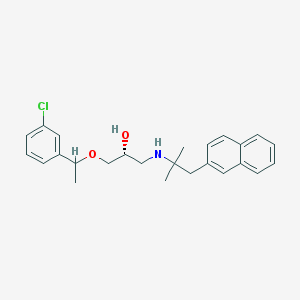 CHEMBL1083308 CHEMBL1083308 | C25H30ClNO2 | 411.97 | 3 / 2 | 5.4 | No |
| 56450 | 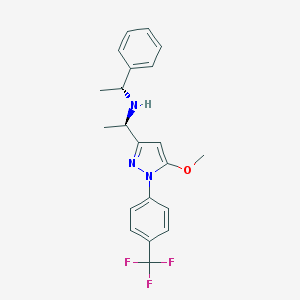 CHEMBL569863 CHEMBL569863 | C21H22F3N3O | 389.422 | 6 / 1 | 4.8 | Yes |
| 523248 |  CHEMBL3827898 CHEMBL3827898 | C28H35NO7S | 529.648 | 8 / 3 | 0.0 | No |
| 58809 | 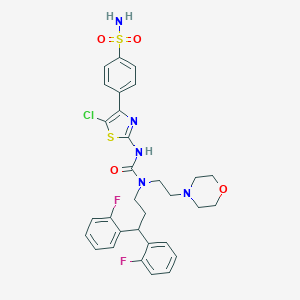 CHEMBL3093425 CHEMBL3093425 | C31H32ClF2N5O4S2 | 676.195 | 10 / 2 | 5.3 | No |
| 523264 | 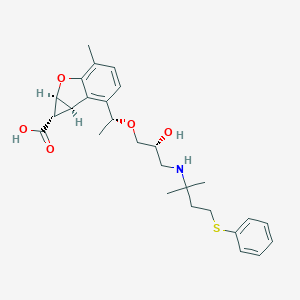 CHEMBL3827577 CHEMBL3827577 | C27H35NO5S | 485.639 | 7 / 3 | 1.4 | Yes |
| 62284 |  CHEMBL1801171 CHEMBL1801171 | C21H23ClN2O3S | 418.936 | 5 / 3 | N/A | N/A |
| 63733 | 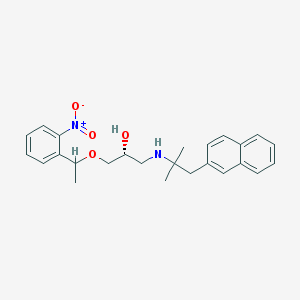 CHEMBL1083314 CHEMBL1083314 | C25H30N2O4 | 422.525 | 5 / 2 | 4.6 | Yes |
| 64080 |  CHEMBL1084514 CHEMBL1084514 | C28H36N2O4 | 464.606 | 6 / 2 | 4.2 | Yes |
| 64896 | 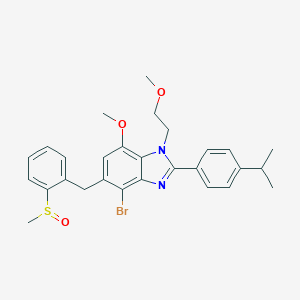 CHEMBL1222492 CHEMBL1222492 | C28H31BrN2O3S | 555.531 | 5 / 0 | 5.7 | No |
| 65676 | 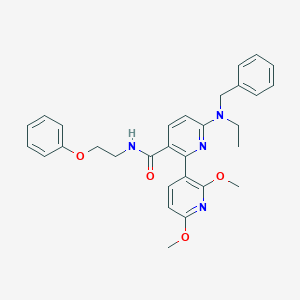 CHEMBL455406 CHEMBL455406 | C30H32N4O4 | 512.61 | 7 / 1 | 5.3 | No |
| 537572 | 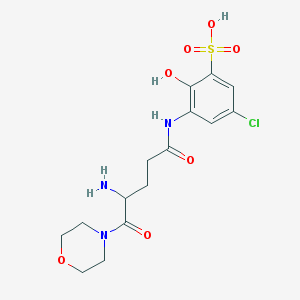 CHEMBL3949209 CHEMBL3949209 | C15H20ClN3O7S | 421.849 | 8 / 4 | -3.1 | Yes |
| 65973 | 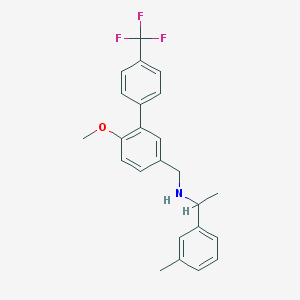 CHEMBL1224343 CHEMBL1224343 | C24H24F3NO | 399.457 | 5 / 1 | 6.0 | No |
| 559428 | 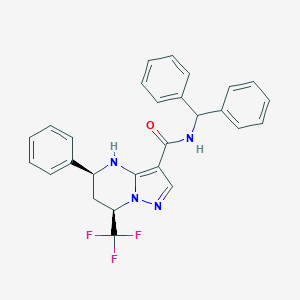 CHEMBL1688102 CHEMBL1688102 | C27H23F3N4O | 476.503 | 6 / 2 | 5.8 | No |
| 68650 | 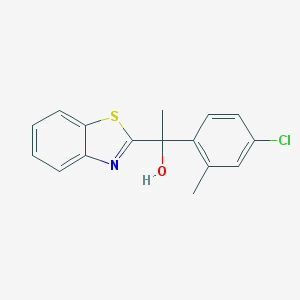 CHEMBL1256403 CHEMBL1256403 | C16H14ClNOS | 303.804 | 3 / 1 | 4.4 | Yes |
| 68842 | 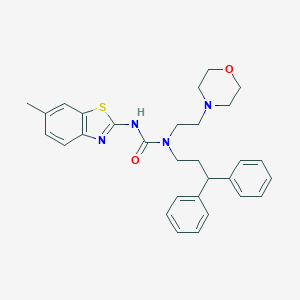 CHEMBL2377749 CHEMBL2377749 | C30H34N4O2S | 514.688 | 5 / 1 | 5.7 | No |
| 537668 | 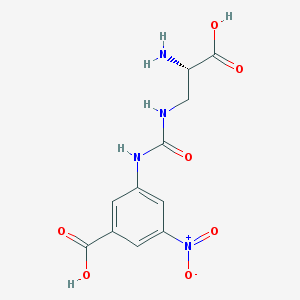 CHEMBL3946636 CHEMBL3946636 | C11H12N4O7 | 312.238 | 8 / 5 | -3.3 | Yes |
| 70423 | 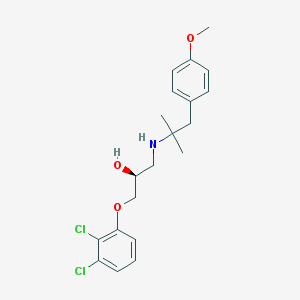 CHEMBL487508 CHEMBL487508 | C20H25Cl2NO3 | 398.324 | 4 / 2 | 4.6 | Yes |
| 70426 | 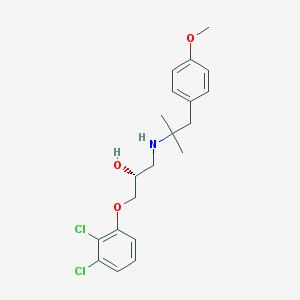 CHEMBL487507 CHEMBL487507 | C20H25Cl2NO3 | 398.324 | 4 / 2 | 4.6 | Yes |
| 70428 | 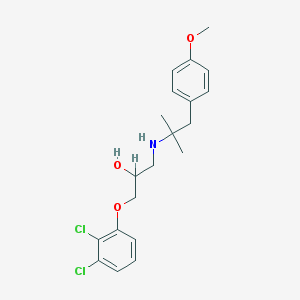 CHEMBL491251 CHEMBL491251 | C20H25Cl2NO3 | 398.324 | 4 / 2 | 4.6 | Yes |
| 70836 |  CHEMBL2349582 CHEMBL2349582 | C35H45N3O4 | 571.762 | 5 / 1 | 6.6 | No |
| 71039 | 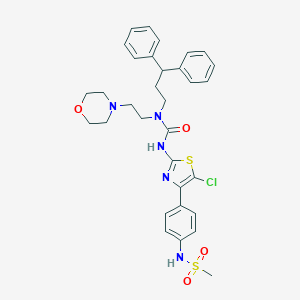 CHEMBL3093421 CHEMBL3093421 | C32H36ClN5O4S2 | 654.241 | 8 / 2 | 5.5 | No |
| 72286 | 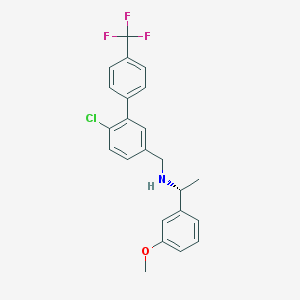 CHEMBL1224344 CHEMBL1224344 | C23H21ClF3NO | 419.872 | 5 / 1 | 6.3 | No |
| 72306 | 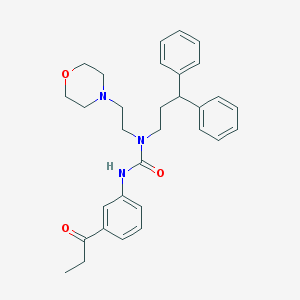 CHEMBL2349586 CHEMBL2349586 | C31H37N3O3 | 499.655 | 4 / 1 | 4.7 | Yes |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218