

You can:
| Name | METHDILAZINE |
|---|---|
| Molecular formula | C18H20N2S |
| IUPAC name | 10-[(1-methylpyrrolidin-3-yl)methyl]phenothiazine |
| Molecular weight | 296.432 |
| Hydrogen bond acceptor | 3 |
| Hydrogen bond donor | 0 |
| XlogP | 5.2 |
| Synonyms | Disyncran (Salt/Mix) Methdilazine (INN) Metodilazina [INN-Spanish] PDSP2_000148 Tacaryl (TN) [ Show all ] |
| Inchi Key | HTMIBDQKFHUPSX-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C18H20N2S/c1-19-11-10-14(12-19)13-20-15-6-2-4-8-17(15)21-18-9-5-3-7-16(18)20/h2-9,14H,10-13H2,1H3 |
| PubChem CID | 14677 |
| ChEMBL | N/A |
| IUPHAR | N/A |
| BindingDB | 81470 |
| DrugBank | DB00902 |
Structure | 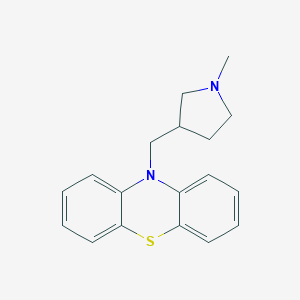 |
| Lipinski's druglikeness | This ligand has a partition coefficient log P greater than 5. |
You can:
| GLASS ID | Name | UniProt | Gene | Species | Length |
|---|---|---|---|---|---|
| 123624 | Histamine H1 receptor | P35367 | HRH1 | Homo sapiens (Human) | 487 |
| 123625 | Histamine H1 receptor | P31390 | Hrh1 | Rattus norvegicus (Rat) | 486 |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218