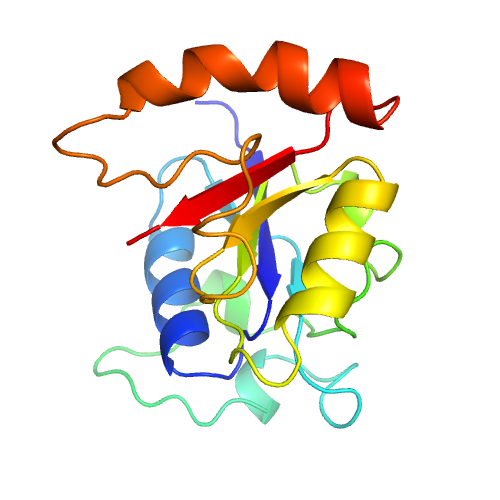
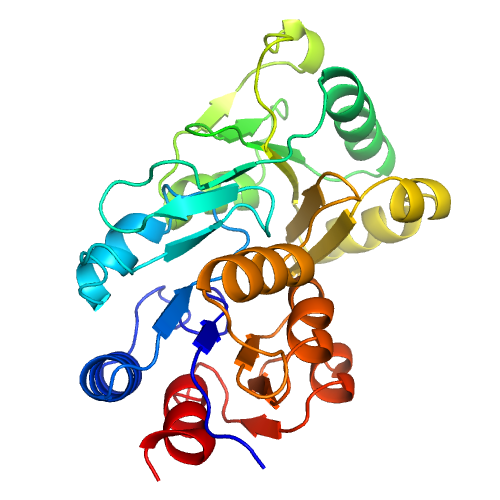
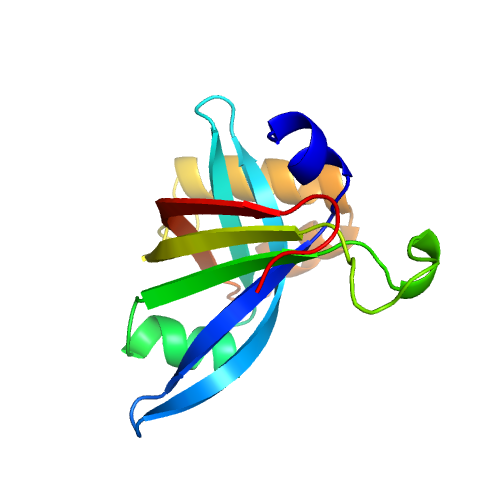
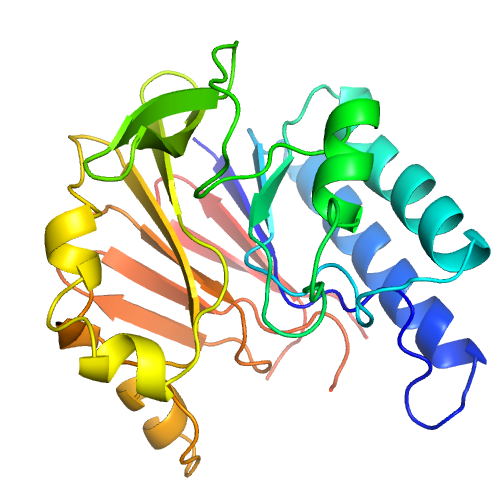
| Rank | PDB Hit | ID1 | ID2 | Cov | Norm. Zscore | DownloadAlignment | 20 40 60 80 100 120 140 160 | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str. Sol.Acc. Seq | CHHHHHHHHHHHCCCCCCSSSCCCCCCCCCCCCHHHHHHHHHHCCCCCCSSSCCCCCCHHHHHHHHHHHCCCCCCCHHHCCSSSSCCCCCCCCCCCCCCSSSSSSCCCCCCCCSSSCCCCCCCCCCCHHHHHHHHHHCCCCCCCSSSSSSSCCSSSSSCCCCSSSSC 85214102300400431433506643643430210000000000145030203552537302520322140432643640220003454425413323000000004517657040423433414036401520472354100000000004520000112141458 QDAQHSFRRLLKASEPGVIVALHQLKRGWQPLNIATTSVLLTLADNDTPVWLSTPLNNDIVNQSLRFHTNAPLVSQPEQATFAVTDEAISSEQLNALSGATLILQVASLSGGRLRLTGEERIAPQLPECILHELTERPHPFPLGIDLILTCGERLLAIPRTTHVEVC | |||||||||||||||||||||||||
| 1 | 4xb6D | 0.16 | 0.16 | 0.96 | 2.29 | CEthreader | QTKRMIRRAILKAVIPGYQVPFGREMPMPYGWGTGGIQLTASVIGESDVLKVIDQGADVSIRNFFKRVTGVNTTERTDDATVIQTRHRIPETPLT--EDQIIIFQVPIPYAYPVKVNGRYVMDPSPIPKFDNPKMDMMPA-----LQLFGAGRRIYAVPPFTRVESL | |||||||||||||||||||
| 2 | 4xb6D | 0.17 | 0.16 | 0.93 | 0.81 | SparksX | QTKRMIRRAILKAAIPGYQVPFGGREMPMPYWGTGGIQLTASVIGESDVLKVIDQGADDTIRNFFKRVTGVNTTERTDDATVIQTRH-----RIPETPLQIIIFQVPIIATTYVKVNGRYVMDPPIPKFDNPKMDMMPAL------QLFGAGRRIYAVPPFTRVESL | |||||||||||||||||||
| 3 | 4xb6D | 0.16 | 0.15 | 0.94 | 0.46 | HHpred | -TKRMIRRAILKAAIPGYQVPFGGRMPMPYGWGTGGIQLTASVIGESDVLKIDQGADDVSIRNFFKRVTGVNTTERTDDATV--IQTRHRIPE-TPLTDQIIIFQVPIPEPGHVKVNGRYVMPSPIPKFDNPKMDMMPAL------QLFGAGKRIYAVPPFTRVESL | |||||||||||||||||||
| 4 | 4xb6D | 0.16 | 0.16 | 0.95 | 0.65 | FFAS3D | --KRMIRRAILKAAIPGYQVPFGGREMPMPGWGTGGIQLTASVIGESDVLKVDDTTNAVSIRNFFKRVTGVNTTERTDDATVIQTRHRIPETPL--TEDQIIIFQVPTTYAYPVKVNGRYVMDPSPIPKFDNPKMDMMPAL-----QLFGAGRRIYAVPPFTRVESL | |||||||||||||||||||
| 5 | 5cefA1 | 0.14 | 0.11 | 0.79 | 0.64 | MUSTER | TVGQRFIELLAAH--PYALHALGASSRSAGQQYARVV-------------WKLPSPIPDAVRHM-VVHECRPDAPGFAECGVVFSDADVAGDIENAFADLVVYSNAKNYRRDP----LCPLIVPLVNPSHLSIIPYQREQ--LGLK-----KGYIVTNA-------- | |||||||||||||||||||
| 6 | 1rkqA | 0.12 | 0.12 | 1.00 | 0.69 | Neff-MUSTER | PAVKNAIAAARARPYAGVHNYLKELHMEQPGDYCITYNGALVQKAADGSTVAQTALSYDDYRFLEKLSREVGSHFHALDRTTLYTANRDISYYTVHESVATIPLVFCEAEKMDPNTQFLKVMMIDEPAILDQAIARIPQEVKEKYTVLKSAPYFLEILDKRVNKGTG | |||||||||||||||||||
| 7 | 1vl7A | 0.07 | 0.05 | 0.76 | 0.53 | PPAS | ------YAGFIQEFQSAIISTISEQGIP--------GSYAPFVIDDAKNIYIYVSGLAVHTKNIEA-----------PLVNVLFVDDEAKTN--------------QIFARRRLSFDCTATLIERESQKWNQVVDQFQERFGQIIEVLRGLADRIFQL-TPKEGRFV | |||||||||||||||||||
| 8 | 4xb6D | 0.17 | 0.16 | 0.93 | 0.61 | HHsearch | QTKRMIRRAILKAAIPGYQVPFGGR-EMPYGWGTGGIQLTASVIGESDVLKVIDQGADVSIRNFFKRVTGVNTTERTDDATVIQTRHRIPETP---LTDQIIIFQVPIPEPLYVKVNGRYVMPSPIPK--FDNPKMDMMPA---L-QLFGAGRRIYAVPPFTRVES- | |||||||||||||||||||
| 9 | 4heqA | 0.13 | 0.10 | 0.74 | 0.70 | SP3 | EGAEAIAKTLNSE---GETTVVNVADVTAPGLAEGYDVVLLG-----CSTWGDDEIEQEDFVPLYEDLDRAGLKDKK-VGVFGCGDSSYTYFCGAVDV-----IE-KKAEELGATLVASSLKIDGEPLDWAREVLARV----------------------------- | |||||||||||||||||||
| 10 | 1s8eA1 | 0.12 | 0.11 | 0.95 | 0.62 | PROSPECTOR2 | MEFAEAFKNALEQENVDFILILFHSSRPSPGTLKKAIALLQIPKEHSIPVFAVRTQRGPSVLNLLEDFGLVYVIEKVENEYLTSERLGNGEYLVKGVYKDLEIHGMKYMSSAW---------FEANKEILVREVSEARGELPEGYLYYALYSGSPVVYPGSLKPRFV | |||||||||||||||||||
| ||||||||||||||||||||||||||
| ||||||||||||||||||||||||||