

You can:
| Name | Orexin receptor type 1 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | HCRTR1 |
| Synonym | hypocretin receptor 1 OX1R Ox1-R OX1 receptor Ox-1-R [ Show all ] |
| Disease | Insomnia Sleep disorders; Insomnia; Substance dependence Sleep disorders; Insomnia |
| Length | 425 |
| Amino acid sequence | MEPSATPGAQMGVPPGSREPSPVPPDYEDEFLRYLWRDYLYPKQYEWVLIAAYVAVFVVALVGNTLVCLAVWRNHHMRTVTNYFIVNLSLADVLVTAICLPASLLVDITESWLFGHALCKVIPYLQAVSVSVAVLTLSFIALDRWYAICHPLLFKSTARRARGSILGIWAVSLAIMVPQAAVMECSSVLPELANRTRLFSVCDERWADDLYPKIYHSCFFIVTYLAPLGLMAMAYFQIFRKLWGRQIPGTTSALVRNWKRPSDQLGDLEQGLSGEPQPRARAFLAEVKQMRARRKTAKMLMVVLLVFALCYLPISVLNVLKRVFGMFRQASDREAVYACFTFSHWLVYANSAANPIIYNFLSGKFREQFKAAFSCCLPGLGPCGSLKAPSPRSSASHKSLSLQSRCSISKISEHVVLTSVTTVLP |
| UniProt | O43613 |
| Protein Data Bank | 4zjc, 4zj8 |
| GPCR-HGmod model | O43613 |
| 3D structure model | This structure is from PDB ID 4zjc. |
| BioLiP | BL0339163, BL0339164 |
| Therapeutic Target Database | T73482 |
| ChEMBL | CHEMBL5113 |
| IUPHAR | 321 |
| DrugBank | BE0005864 |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 57 |  CID 44405017 CID 44405017 | C128H211N45O35S | 2972.44 | 41 / 42 | -12.2 | No |
| 202 |  SCHEMBL1671891 SCHEMBL1671891 | C27H24N4OS | 452.576 | 5 / 0 | 4.9 | Yes |
| 356 |  CHEMBL3667557 CHEMBL3667557 | C18H19ClN2O3S2 | 410.931 | 5 / 1 | 3.6 | Yes |
| 358 |  SCHEMBL2026090 SCHEMBL2026090 | C26H27FN4O3 | 462.525 | 7 / 0 | 3.0 | Yes |
| 389 | 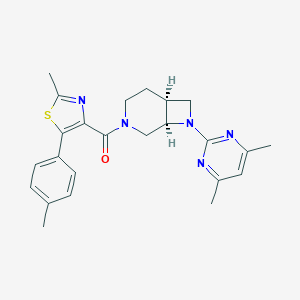 CHEMBL3670580 CHEMBL3670580 | C24H27N5OS | 433.574 | 6 / 0 | 4.6 | Yes |
| 463032 |  CHEMBL3663396 CHEMBL3663396 | C25H26N2O3 | 402.494 | 4 / 0 | 4.9 | Yes |
| 729 | 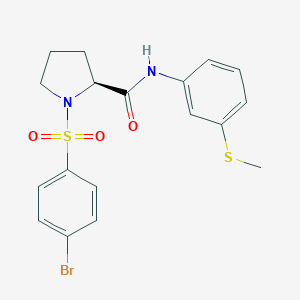 CHEMBL3667561 CHEMBL3667561 | C18H19BrN2O3S2 | 455.385 | 5 / 1 | 3.7 | Yes |
| 517321 | 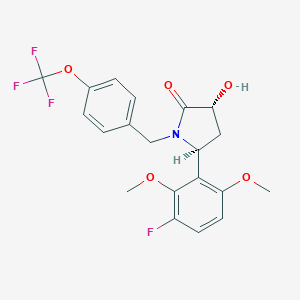 SCHEMBL4536874 SCHEMBL4536874 | C20H19F4NO5 | 429.368 | 9 / 1 | 3.5 | Yes |
| 876 | 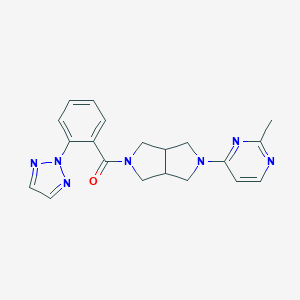 SCHEMBL1671846 SCHEMBL1671846 | C20H21N7O | 375.436 | 6 / 0 | 2.3 | Yes |
| 557331 | 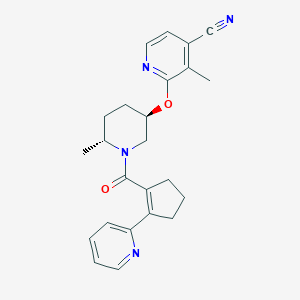 SCHEMBL17043222 SCHEMBL17043222 | C24H26N4O2 | 402.498 | 5 / 0 | 3.2 | Yes |
| 926 |  SCHEMBL6924756 SCHEMBL6924756 | C26H25N7O | 451.534 | 6 / 0 | 3.9 | Yes |
| 535941 | 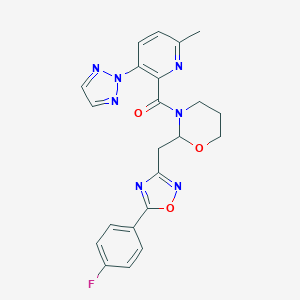 CHEMBL3893150 CHEMBL3893150 | C22H20FN7O3 | 449.446 | 9 / 0 | 3.1 | Yes |
| 1145 | 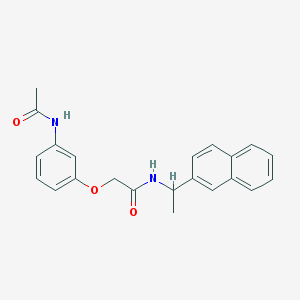 MLS001177268 MLS001177268 | C22H22N2O3 | 362.429 | 3 / 2 | 3.6 | Yes |
| 1240 | 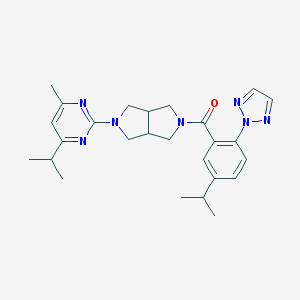 SCHEMBL1671969 SCHEMBL1671969 | C26H33N7O | 459.598 | 6 / 0 | 4.5 | Yes |
| 1289 | 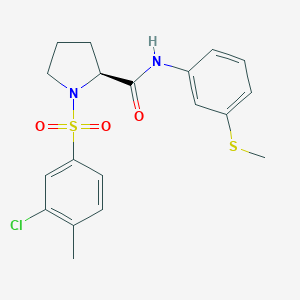 CHEMBL3667560 CHEMBL3667560 | C19H21ClN2O3S2 | 424.958 | 5 / 1 | 4.0 | Yes |
| 463140 |  CHEMBL3659195 CHEMBL3659195 | C22H21FN4O2 | 392.434 | 5 / 0 | 3.4 | Yes |
| 1641 | 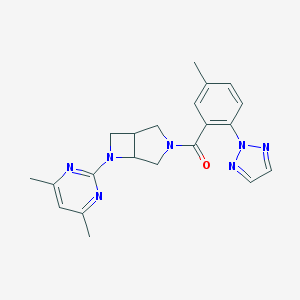 SCHEMBL1671936 SCHEMBL1671936 | C21H23N7O | 389.463 | 6 / 0 | 2.8 | Yes |
| 459246 | 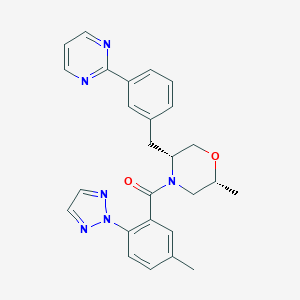 CHEMBL3894723 CHEMBL3894723 | C26H26N6O2 | 454.534 | 6 / 0 | 3.8 | Yes |
| 1731 | 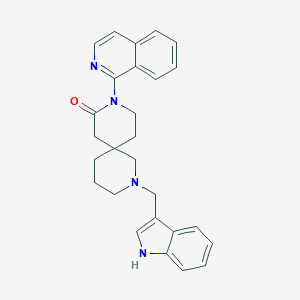 CHEMBL3669566 CHEMBL3669566 | C27H28N4O | 424.548 | 3 / 1 | 4.1 | Yes |
| 1833 | 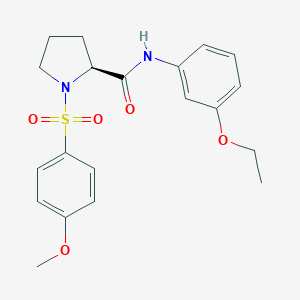 CHEMBL3667579 CHEMBL3667579 | C20H24N2O5S | 404.481 | 6 / 1 | 2.8 | Yes |
| 535961 | 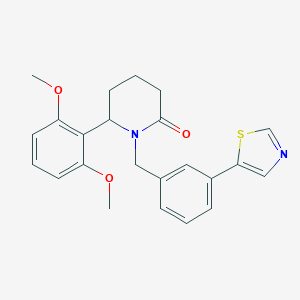 SCHEMBL4536732 SCHEMBL4536732 | C23H24N2O3S | 408.516 | 5 / 0 | 3.8 | Yes |
| 2027 |  SCHEMBL1671105 SCHEMBL1671105 | C27H23FN4O | 438.506 | 5 / 0 | 4.6 | Yes |
| 533919 | 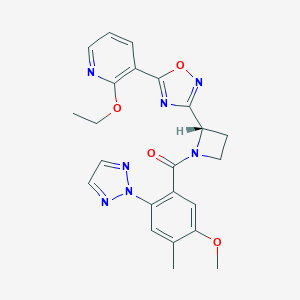 CHEMBL3970835 CHEMBL3970835 | C23H23N7O4 | 461.482 | 9 / 0 | 2.9 | Yes |
| 2110 | 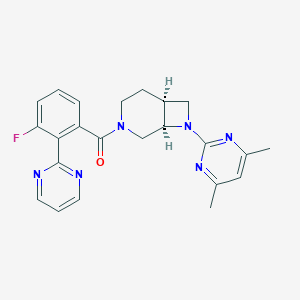 SCHEMBL1671801 SCHEMBL1671801 | C23H23FN6O | 418.476 | 7 / 0 | 2.9 | Yes |
| 2394 | 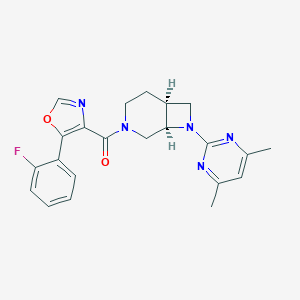 SCHEMBL1673234 SCHEMBL1673234 | C22H22FN5O2 | 407.449 | 7 / 0 | 3.4 | Yes |
| 517330 |  SCHEMBL4536467 SCHEMBL4536467 | C23H24N2O3 | 376.456 | 4 / 0 | 3.4 | Yes |
| 517332 | 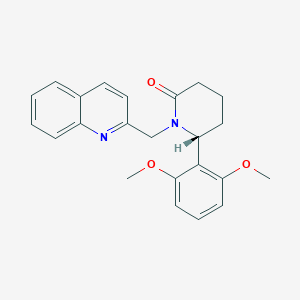 SCHEMBL4534799 SCHEMBL4534799 | C23H24N2O3 | 376.456 | 4 / 0 | 3.4 | Yes |
| 2422 |  CHEMBL3112601 CHEMBL3112601 | C23H24N2O3 | 376.456 | 4 / 0 | 3.4 | Yes |
| 535999 | 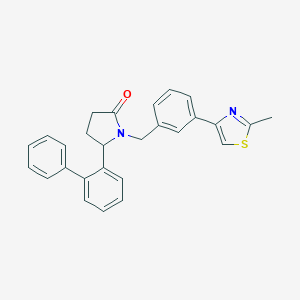 SCHEMBL4529392 SCHEMBL4529392 | C27H24N2OS | 424.562 | 3 / 0 | 5.5 | No |
| 3458 | 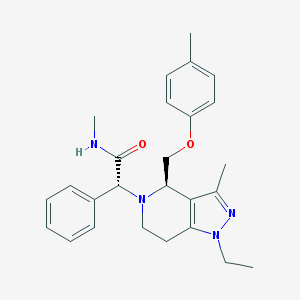 CHEMBL1080571 CHEMBL1080571 | C26H32N4O2 | 432.568 | 4 / 1 | 3.9 | Yes |
| 459255 |  CHEMBL3909223 CHEMBL3909223 | C26H25FN6O2 | 472.524 | 7 / 0 | 3.9 | Yes |
| 3514 |  CHEMBL3235265 CHEMBL3235265 | C25H26N4O2 | 414.509 | 4 / 1 | 3.9 | Yes |
| 3601 |  SMR000010089 SMR000010089 | C20H20N2O3 | 336.391 | 4 / 1 | 3.6 | Yes |
| 536012 |  SCHEMBL16464778 SCHEMBL16464778 | C24H24N6O4 | 460.494 | 8 / 0 | 3.6 | Yes |
| 3785 | 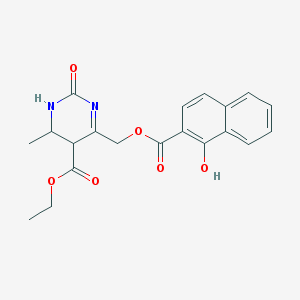 BDBM79952 BDBM79952 | C20H20N2O6 | 384.388 | 6 / 2 | 3.4 | Yes |
| 557387 | 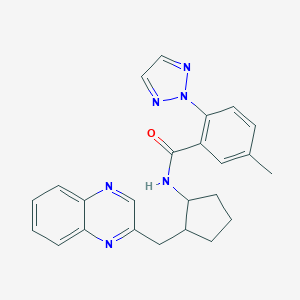 SCHEMBL16655666 SCHEMBL16655666 | C24H24N6O | 412.497 | 5 / 1 | 4.0 | Yes |
| 536016 | 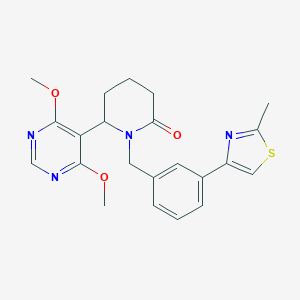 SCHEMBL4541596 SCHEMBL4541596 | C22H24N4O3S | 424.519 | 7 / 0 | 3.2 | Yes |
| 521552 | 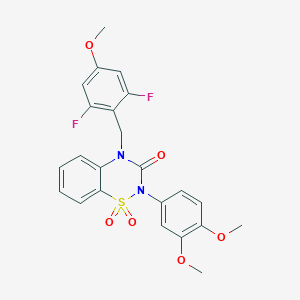 CHEMBL3742398 CHEMBL3742398 | C23H20F2N2O6S | 490.478 | 8 / 0 | 3.7 | Yes |
| 536019 | 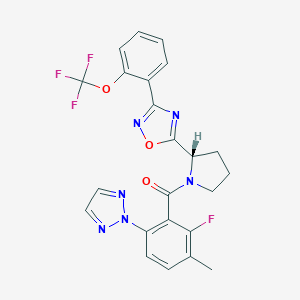 SCHEMBL16463160 SCHEMBL16463160 | C23H18F4N6O3 | 502.43 | 11 / 0 | 4.9 | No |
| 3977 | 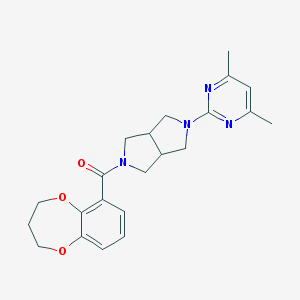 SCHEMBL1672410 SCHEMBL1672410 | C22H26N4O3 | 394.475 | 6 / 0 | 2.8 | Yes |
| 536021 | 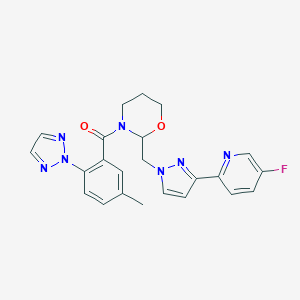 CHEMBL3985942 CHEMBL3985942 | C23H22FN7O2 | 447.474 | 7 / 0 | 2.5 | Yes |
| 536024 | 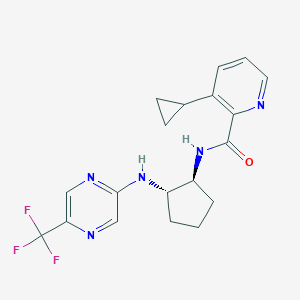 CHEMBL3965933 CHEMBL3965933 | C19H20F3N5O | 391.398 | 8 / 2 | 3.1 | Yes |
| 4083 | 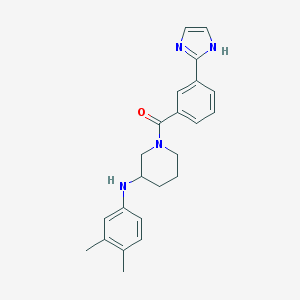 MLS001072368 MLS001072368 | C23H26N4O | 374.488 | 3 / 2 | 4.1 | Yes |
| 4255 |  MLS001223988 MLS001223988 | C22H28N2O4S | 416.536 | 5 / 1 | 3.5 | Yes |
| 536038 | 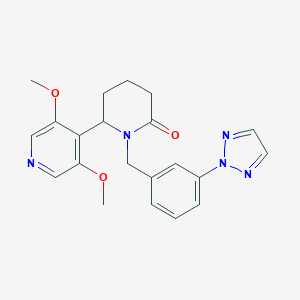 SCHEMBL4532006 SCHEMBL4532006 | C21H23N5O3 | 393.447 | 6 / 0 | 2.0 | Yes |
| 517342 | 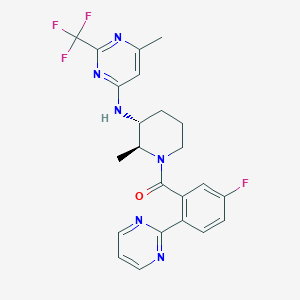 CHEMBL3704932 CHEMBL3704932 | C23H22F4N6O | 474.464 | 10 / 1 | 4.0 | Yes |
| 4432 | 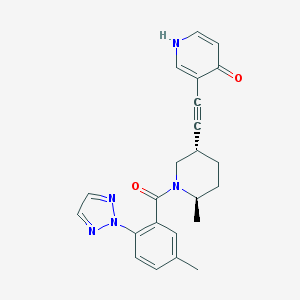 CHEMBL3235258 CHEMBL3235258 | C23H23N5O2 | 401.47 | 5 / 1 | 3.0 | Yes |
| 4494 | 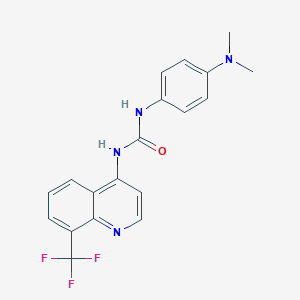 CHEMBL1771432 CHEMBL1771432 | C19H17F3N4O | 374.367 | 6 / 2 | 3.8 | Yes |
| 536053 | 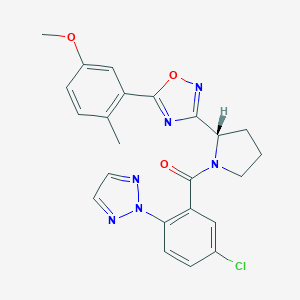 SCHEMBL16463647 SCHEMBL16463647 | C23H21ClN6O3 | 464.91 | 7 / 0 | 4.3 | Yes |
| 463464 | 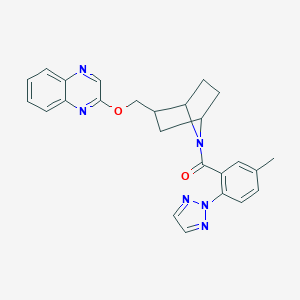 CHEMBL3659205 CHEMBL3659205 | C25H24N6O2 | 440.507 | 6 / 0 | 4.0 | Yes |
| 536056 |  SCHEMBL4540837 SCHEMBL4540837 | C25H25FN2O3 | 420.484 | 5 / 0 | 3.8 | Yes |
| 4898 | 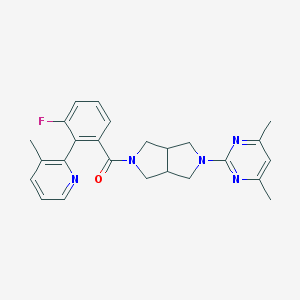 SCHEMBL6925282 SCHEMBL6925282 | C25H26FN5O | 431.515 | 6 / 0 | 3.7 | Yes |
| 557416 | 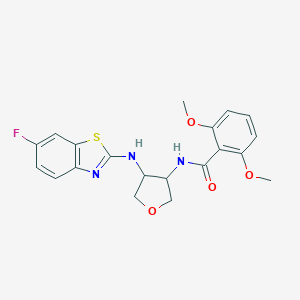 SCHEMBL16666146 SCHEMBL16666146 | C20H20FN3O4S | 417.455 | 8 / 2 | 3.5 | Yes |
| 5138 | 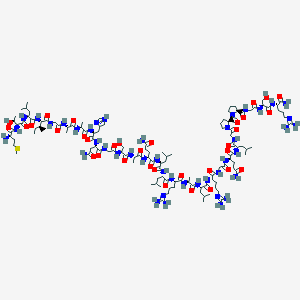 CID 44404999 CID 44404999 | C121H209N43O34S | 2842.33 | 40 / 40 | -11.4 | No |
| 521614 | 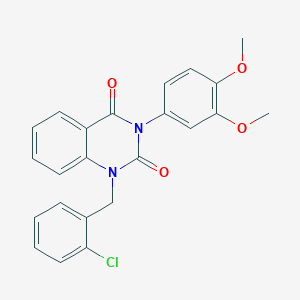 CHEMBL3741065 CHEMBL3741065 | C23H19ClN2O4 | 422.865 | 4 / 0 | 4.4 | Yes |
| 536074 | 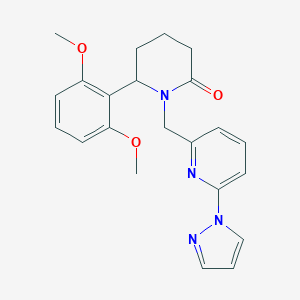 SCHEMBL4536986 SCHEMBL4536986 | C22H24N4O3 | 392.459 | 5 / 0 | 2.4 | Yes |
| 5457 | 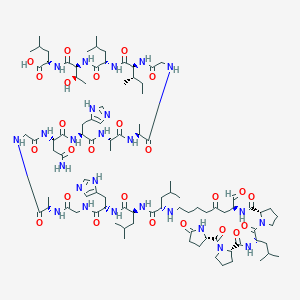 BDBM50140022 BDBM50140022 | C94H151N25O25 | 2031.39 | 28 / 23 | -3.4 | No |
| 5525 | 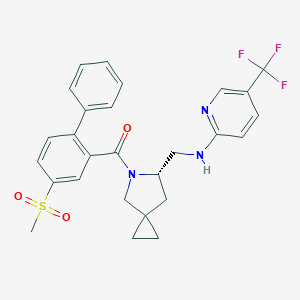 CHEMBL2381082 CHEMBL2381082 | C27H26F3N3O3S | 529.578 | 8 / 1 | 4.9 | No |
| 5692 | 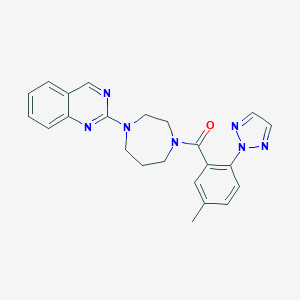 CHEMBL469146 CHEMBL469146 | C23H23N7O | 413.485 | 6 / 0 | 3.6 | Yes |
| 459272 | 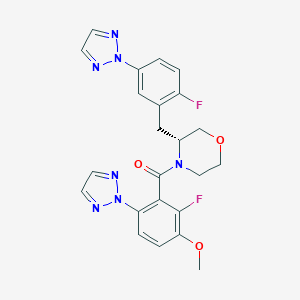 CHEMBL3974582 CHEMBL3974582 | C23H21F2N7O3 | 481.464 | 9 / 0 | 3.2 | Yes |
| 6112 |  SCHEMBL2027169 SCHEMBL2027169 | C25H26N6O | 426.524 | 5 / 1 | 2.5 | Yes |
| 6292 | 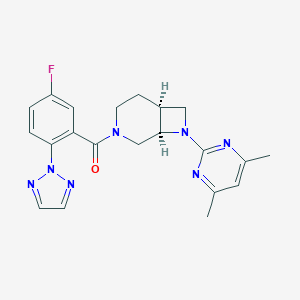 CHEMBL2413512 CHEMBL2413512 | C21H22FN7O | 407.453 | 7 / 0 | 2.9 | Yes |
| 6294 | 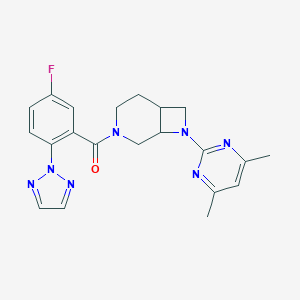 CHEMBL3670630 CHEMBL3670630 | C21H22FN7O | 407.453 | 7 / 0 | 2.9 | Yes |
| 536098 |  SCHEMBL16465942 SCHEMBL16465942 | C23H21ClN6O3 | 464.91 | 7 / 0 | 4.3 | Yes |
| 6320 | 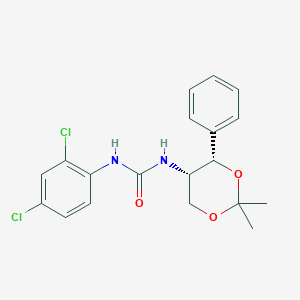 CHEMBL185136 CHEMBL185136 | C19H20Cl2N2O3 | 395.28 | 3 / 2 | 4.0 | Yes |
| 517352 |  SCHEMBL4536325 SCHEMBL4536325 | C20H18ClF4NO3 | 431.812 | 7 / 0 | 5.4 | No |
| 463640 |  CHEMBL3622428 CHEMBL3622428 | C32H34N4O6S | 602.706 | 8 / 3 | 4.4 | No |
| 6443 | 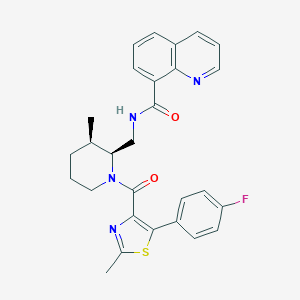 CHEMBL2031484 CHEMBL2031484 | C28H27FN4O2S | 502.608 | 6 / 1 | 5.5 | No |
| 547967 | 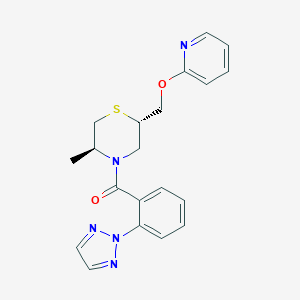 CHEMBL3901510 CHEMBL3901510 | C20H21N5O2S | 395.481 | 6 / 0 | 3.2 | Yes |
| 557461 | 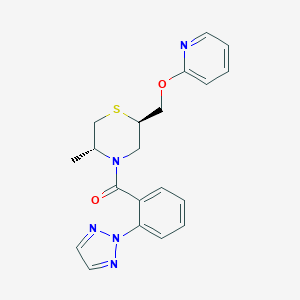 SCHEMBL16033642 SCHEMBL16033642 | C20H21N5O2S | 395.481 | 6 / 0 | 3.2 | Yes |
| 6549 | 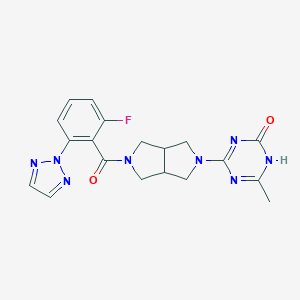 CHEMBL3649206 CHEMBL3649206 | C19H19FN8O2 | 410.413 | 5 / 1 | 0.6 | Yes |
| 6624 | 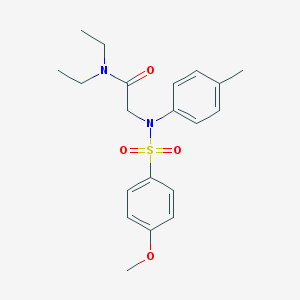 CHEMBL524440 CHEMBL524440 | C20H26N2O4S | 390.498 | 5 / 0 | 3.2 | Yes |
| 557471 |  SCHEMBL16446669 SCHEMBL16446669 | C21H26N6O2S | 426.539 | 7 / 1 | 3.1 | Yes |
| 6633 |  CHEMBL2413372 CHEMBL2413372 | C24H22FN3O3S | 451.516 | 5 / 0 | 3.9 | Yes |
| 521642 |  CHEMBL3741474 CHEMBL3741474 | C22H19F2N3O6S | 491.466 | 9 / 0 | 2.9 | Yes |
| 6696 |  CHEMBL182269 CHEMBL182269 | C24H25N3O3 | 403.482 | 4 / 2 | 3.3 | Yes |
| 517355 | 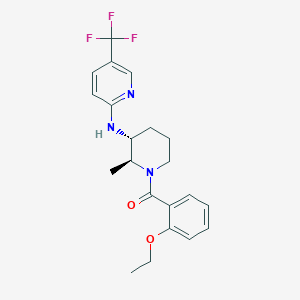 CHEMBL3704949 CHEMBL3704949 | C21H24F3N3O2 | 407.437 | 7 / 1 | 4.5 | Yes |
| 6863 | 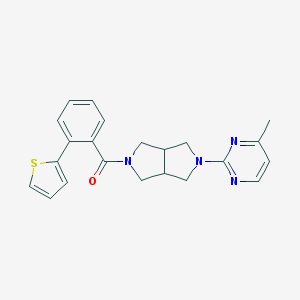 SCHEMBL1671430 SCHEMBL1671430 | C22H22N4OS | 390.505 | 5 / 0 | 3.6 | Yes |
| 6873 | 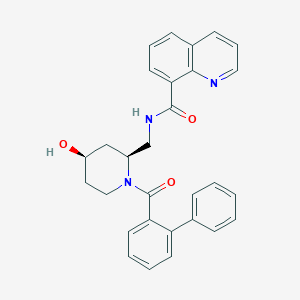 CHEMBL2031494 CHEMBL2031494 | C29H27N3O3 | 465.553 | 4 / 2 | 4.2 | Yes |
| 536115 |  SCHEMBL4536426 SCHEMBL4536426 | C25H26N2O3 | 402.494 | 4 / 0 | 4.0 | Yes |
| 7061 | 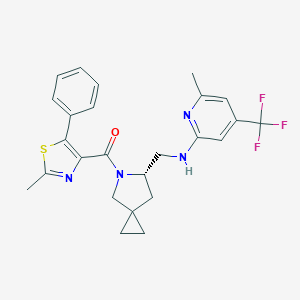 CHEMBL2381089 CHEMBL2381089 | C25H25F3N4OS | 486.557 | 8 / 1 | 5.8 | No |
| 557484 | 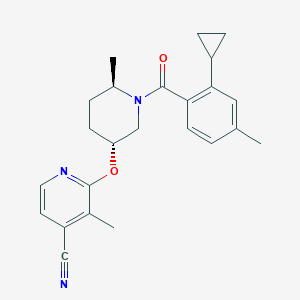 SCHEMBL15658462 SCHEMBL15658462 | C24H27N3O2 | 389.499 | 4 / 0 | 4.7 | Yes |
| 536155 | 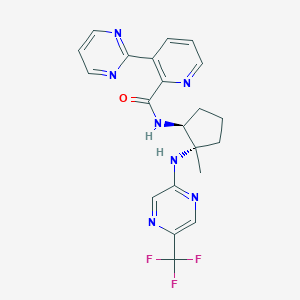 CHEMBL3910747 CHEMBL3910747 | C21H20F3N7O | 443.434 | 10 / 2 | 2.4 | Yes |
| 441989 | 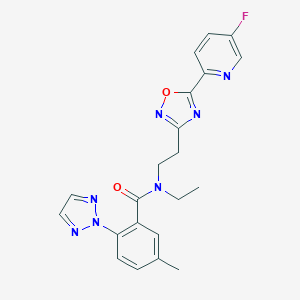 CHEMBL3398471 CHEMBL3398471 | C21H20FN7O2 | 421.436 | 8 / 0 | 3.0 | Yes |
| 463808 | 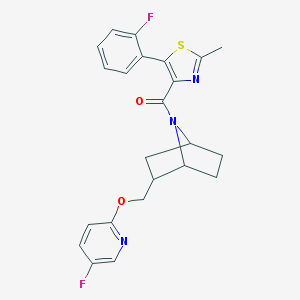 CHEMBL3659198 CHEMBL3659198 | C23H21F2N3O2S | 441.497 | 7 / 0 | 4.9 | Yes |
| 536165 | 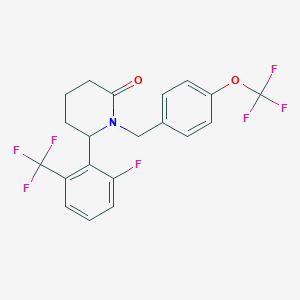 SCHEMBL4534316 SCHEMBL4534316 | C20H16F7NO2 | 435.342 | 9 / 0 | 5.3 | No |
| 7639 | 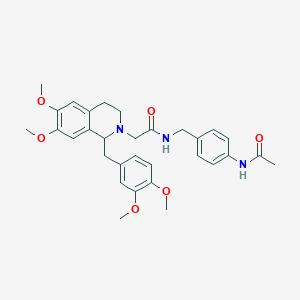 CHEMBL2418838 CHEMBL2418838 | C31H37N3O6 | 547.652 | 7 / 2 | 3.7 | No |
| 536170 | 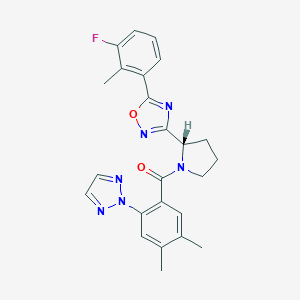 SCHEMBL16465387 SCHEMBL16465387 | C24H23FN6O2 | 446.486 | 7 / 0 | 4.5 | Yes |
| 7695 | 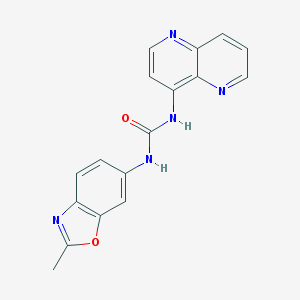 792173-99-0 792173-99-0 | C17H13N5O2 | 319.324 | 5 / 2 | 2.0 | Yes |
| 7740 |  CHEMBL1272096 CHEMBL1272096 | C17H15N5O3S2 | 401.459 | 8 / 1 | 2.4 | Yes |
| 7904 |  MLS000096514 MLS000096514 | C16H11NO4S2 | 345.387 | 7 / 0 | 4.0 | Yes |
| 459288 |  CHEMBL3896431 CHEMBL3896431 | C24H21ClN6O2 | 460.922 | 6 / 0 | 3.6 | Yes |
| 8107 |  SCHEMBL1671269 SCHEMBL1671269 | C26H23N5O | 421.504 | 5 / 0 | 3.4 | Yes |
| 536179 |  SCHEMBL16464245 SCHEMBL16464245 | C24H23FN6O3 | 462.485 | 8 / 0 | 4.1 | Yes |
| 8260 |  CHEMBL3113758 CHEMBL3113758 | C21H23F3N2O4 | 424.42 | 8 / 0 | 3.7 | Yes |
| 517361 |  SCHEMBL4534291 SCHEMBL4534291 | C20H18ClF4NO4 | 447.811 | 8 / 0 | 5.0 | Yes |
| 536188 |  SCHEMBL17064033 SCHEMBL17064033 | C25H25FN6O3 | 476.512 | 8 / 0 | 4.5 | Yes |
| 8471 |  CHEMBL55270 CHEMBL55270 | C17H13N5O2 | 319.324 | 5 / 2 | 2.3 | Yes |
| 463909 |  CHEMBL3585948 CHEMBL3585948 | C22H21ClN4O2 | 408.886 | 5 / 1 | 3.6 | Yes |
| 533944 |  CHEMBL3922646 CHEMBL3922646 | C22H15F3N4O3S | 472.442 | 10 / 0 | 5.2 | No |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218