| # | PDB (Resolution) |
Length | EC number & GO term |
RNAcentral | Rfam | Taxon | PubMed | ATtRACT motif |
Ligand & binding nucleotides |
|---|---|---|---|---|---|---|---|---|---|
| 201 |
2du3:D (2.6Å) |
71 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000080E059 | RF00005 | Archaeoglobus fulgidus 2234 |
17351629 | Assembly1 protein:A:1~534 933~937 |
|
| 202 |
2du4:C (2.8Å) |
71 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000080E059 | RF00005 | Archaeoglobus fulgidus 2234 |
17351629 | Assembly1 protein:A:1~534 933~937 |
|
| 203 |
2du5:D (3.2Å) |
71 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000080E26B | RF00005 | Archaeoglobus fulgidus 2234 |
17351629 | Assembly1 protein:A:1~534 933~937 |
|
| 204 |
2du6:D (3.3Å) |
71 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000080E16C | RF00005 | Archaeoglobus fulgidus 2234 |
17351629 | Assembly1 protein:A:1~534 934~936 |
|
| 205 |
2dxi:C (2.2Å) |
75 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000052BB59 | RF00005 | Thermus thermophilus 274 |
17161369 | Assembly1 protein:A:1~468 503,505,511~514,524~525,534~53 9,569~571,574~576 |
|
| 206 |
2dxi:D (2.2Å) |
75 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000052BB59 | RF00005 | Thermus thermophilus 274 |
17161369 | Assembly2 protein:B:1~468 503~506,511~513,524~525,534~53 8,569~570,574~576 |
|
| 207 |
2ees:A (1.75Å) |
67 | GO:0009113 (P) purine nucleobase biosy... | URS000080DE3D | RF00167 | Bacillus subtilis 1423 |
17960911 | Assembly1 ACT:A:96 32~33,38~39,66 NCO:A:101 66~67 NCO:A:102 38,56 NCO:A:103 39,55~56,68~69 NCO:A:104 30~32,66 NCO:A:105 37~38,57 NCO:A:107 41,53~54 NCO:A:108 47,50 NCO:A:109 24,71 NCO:A:111 32~33 HPA:A:90 21~22,47,51~52,74~75 |
|
| 208 |
2eet:A (1.95Å) |
67 | GO:0009113 (P) purine nucleobase biosy... | URS000080DDBB | RF00167 | Bacillus subtilis 1423 |
17960911 | Assembly1 ACT:A:96 32~33,38~39 NCO:A:101 66~67 NCO:A:102 38~39,56 NCO:A:103 39,56,69 NCO:A:104 30~32,66 NCO:A:105 37~38 NCO:A:107 41,53~54 NCO:A:108 47,50~51 NCO:A:111 32~33 HPA:A:90 21~22,47,51~52,74~75 |
|
| 209 |
2eeu:A (1.95Å) |
67 | GO:0009113 (P) purine nucleobase biosy... | URS000080E2FB | RF00167 | Bacillus subtilis 1423 |
17960911 | Assembly1 ACT:A:96 32~33,38~39 NCO:A:101 28,66~67 NCO:A:102 38~39,56 NCO:A:103 39,56,69 NCO:A:104 31~32,66 NCO:A:105 37~38 NCO:A:107 41,53~54 NCO:A:108 46~47 NCO:A:109 24,71 NCO:A:111 32~33 NCO:A:112 72~73 HPA:A:90 21~22,47,51~52,74~75 |
|
| 210 |
2eev:A (1.95Å) |
67 | GO:0009113 (P) purine nucleobase biosy... | URS000080DFC6 | RF00167 | Bacillus subtilis 1423 |
17960911 | Assembly1 ACT:A:95 32~33,38 NCO:A:101 28,66~67 NCO:A:102 38~39,56 NCO:A:103 39,56,69 NCO:A:104 30~32,66 NCO:A:105 37~38 NCO:A:106 26~27 NCO:A:107 41,53~54 NCO:A:108 46~47 NCO:A:109 24,71 NCO:A:110 21~22 NCO:A:111 32~33 NCO:A:112 72~73 HPA:A:90 21~22,47,51~52,74~75 |
|
| 211 |
2eew:A (2.25Å) |
67 | GO:0009113 (P) purine nucleobase biosy... | URS000080E008 | RF00167 | Bacillus subtilis 1423 |
17960911 | Assembly1 ACT:A:96 32~33,38,66 NCO:A:101 66~67 NCO:A:102 38~39,56 NCO:A:103 39,56,69 NCO:A:104 31~32,66 NCO:A:105 37~38 NCO:A:111 32~33 NCO:A:112 72~73 HPA:A:90 21~22,47,51~52,74~75 |
|
| 212 |
2f88:A |
34 | GO:0000373 (P) Group II intron splicing | URS000080E292 | RF00029 | synthetic construct 32630 |
16428604 | ||
| 213 |
2fdt:A |
36 | GO:0032197 (P) retrotransposition | URS000080DE48 | RF00436 | synthetic construct 32630 |
17000640 | ||
| 214 |
2fk6:R (2.9Å) |
53 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000080DE34 | RF00005 | Bacillus subtilis 1423 |
16518398 | Assembly1 protein:A:1~307 3~4,19,56,61~62,73 protein:A-2:1~307 1,72~73 |
|
| 215 |
2fmt:C (2.8Å) |
77 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS00001BBAFC | RF00005 | synthetic construct 32630 |
9843487 | Assembly1 protein:A:1~314 2,11~13,24~25,69~76 |
|
| 216 |
2fmt:D (2.8Å) |
77 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS00001BBAFC | RF00005 | synthetic construct 32630 |
9843487 | Assembly2 protein:B:1~314 2,11~14,24~25,69~76 |
|
| 217 |
2g9c:A (1.7Å) |
67 | GO:0009113 (P) purine nucleobase biosy... | URS000080DFC3 | RF00167 | Bacillus subtilis 1423 |
17076468 | Assembly1 ACT:A:96 32~33,38~39 NCO:A:101 66~67 NCO:A:102 38~39,56 NCO:A:103 39,56,69 NCO:A:104 30~32,66 NCO:A:105 37~38 NCO:A:106 26~27 NCO:A:107 41,53~54 NCO:A:108 47,50~51 NCO:A:109 24,71 NCO:A:111 32~33 3AY:A:91 21~22,51~52,74~75 |
|
| 218 |
2gcs:B (2.1Å) |
123 | GO:0003824 (F) catalytic activity GO:0010468 (P) regulation of gene expr... |
URS000080DF18 | RF00234 | Caldanaerobacter subterraneus subsp. tengcongensis 119072 |
16990543 | Assembly1 rna:A:0~20 23~37,39~40,58,60~65,67,115~11 7 MG:B:501 36~37 MG:B:503 79,81 MG:B:504 57,59 |
|
| 219 |
2gcv:B (2.1Å) |
123 | GO:0003824 (F) catalytic activity GO:0010468 (P) regulation of gene expr... |
URS000080DF18 | RF00234 | Caldanaerobacter subterraneus subsp. tengcongensis 119072 |
16990543 | Assembly1 rna:A:1~20 23~37,58,60~65,115~117 MES:A:501 65~66 MES:B:502 40,65,67 MG:B:401 36~37 MG:B:403 79,81 MG:B:404 57,59 |
|
| 220 |
2gis:A (2.9Å) |
94 | GO:0010468 (P) regulation of gene expr... | URS000080DF35 | RF00162 | Caldanaerobacter subterraneus subsp. tengcongensis 119072 |
16810258 | Assembly1 MG:A:205 10,63~64 MG:A:206 84~85 IRI:A:202 16~18 IRI:A:203 23,25~28 IRI:A:204 4~5,88 SAM:A:301 7,11,45~47,57~59,88~89 |
|
| 221 |
2gm0:A |
35 | GO:0019074 (P) viral RNA genome packaging | URS000080DE12 | RF00175 | Human immunodeficiency virus 1 11676 |
16603544 | Assembly1 rna:B:36~70 1~35 |
|
| 222 |
2gm0:B |
35 | GO:0019074 (P) viral RNA genome packaging | URS000080DE12 | RF00175 | Human immunodeficiency virus 1 11676 |
16603544 | Assembly1 rna:A:1~35 36~70 |
|
| 223 |
2go5:A (7.4Å) |
127 | GO:0006617 (P) SRP-dependent cotransla... GO:0005786 (C) signal recognition part... |
URS000080DFD3 | RF00017 RF01856 |
Canis sp. 9616 |
16675701 | Assembly1 protein:B:14~120 141~143,147~151,195~198,203~20 4 protein:W:326~434 184~185,192~195,207~208 protein:1:-1~130 231~235 |
|
| 224 |
2h0s:B (2.35Å) |
123 | GO:0003824 (F) catalytic activity GO:0010468 (P) regulation of gene expr... |
URS000080DF18 | RF00234 | Caldanaerobacter subterraneus subsp. tengcongensis 119072 |
16990543 | Assembly1 rna:A:-2~20 23~37,39~40,58,60~67,115~117 MG:B:301 36~37 MG:B:302 79,81 MG:B:303 57,59 |
|
| 225 |
2h0w:B (2.4Å) |
123 | GO:0003824 (F) catalytic activity GO:0010468 (P) regulation of gene expr... |
URS000080DF18 | RF00234 | Caldanaerobacter subterraneus subsp. tengcongensis 119072 |
16990543 | Assembly1 rna:A:1~20 23~37,58,60~65,115~117 MG:B:501 36~37 MG:B:503 79,81 MG:B:504 57,59 MES:B:601 65~66 MES:B:602 40,65,67 |
|
| 226 |
2h0x:B (2.3Å) |
123 | GO:0003824 (F) catalytic activity GO:0010468 (P) regulation of gene expr... |
URS000080DF18 | RF00234 | Caldanaerobacter subterraneus subsp. tengcongensis 119072 |
16990543 | Assembly1 rna:A:0~20 23~37,39~40,58,60~65,67,115~11 7 MG:B:501 36~37 MG:B:503 79,81 MG:B:504 57,59 |
|
| 227 |
2h0z:B (2.7Å) |
123 | GO:0003824 (F) catalytic activity GO:0010468 (P) regulation of gene expr... |
URS000080DF18 | RF00234 | Caldanaerobacter subterraneus subsp. tengcongensis 119072 |
16990543 | Assembly1 rna:A:0~20 23~35,37,39~40,58,60~67,115~11 7 G6P:A:301 51,66 MG:A:401 35~36 MG:B:402 57,59 MG:B:403 79,81 |
|
| 228 |
2ho6:B (2.8Å) |
123 | GO:0003824 (F) catalytic activity GO:0010468 (P) regulation of gene expr... |
URS000080DF18 | RF00234 | Caldanaerobacter subterraneus subsp. tengcongensis 119072 |
16990543 | Assembly1 rna:A:1~20 23~37,58,60~65,115~117 MG:B:201 36~37 MG:B:202 79,81 MG:B:203 57,59 MES:B:402 40,65,67 |
|
| 229 |
2ho7:B (2.9Å) |
123 | GO:0003824 (F) catalytic activity GO:0010468 (P) regulation of gene expr... |
URS000080DF18 | RF00234 | Caldanaerobacter subterraneus subsp. tengcongensis 119072 |
16990543 | Assembly1 rna:A:0~20 23~37,39~40,58,60~67,115~117 G6P:A:401 50,65 MG:B:201 36~37 MG:B:202 57,59 MG:B:204 79,81~82 |
|
| 230 |
2iy3:B (16.0Å) |
110 | GO:0006617 (P) SRP-dependent cotransla... GO:0048501 (C) signal recognition part... |
URS000080DE7C | RF00169 | Escherichia coli 562 |
17086205 | ||
| 231 |
2iy5:T (3.1Å) |
76 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000080E078 | RF00005 | Thermus thermophilus HB8 300852 |
16939209 | Assembly1 protein:A:15~350 19,28,42~45,56~57 protein:B:1~780 1,11~12,24~26,33~37 protein:A-2:15~350 69,76 protein:B-2:1~780 10~11,67,73~75 |
|
| 232 |
2j28:8 (9.5Å) |
74 | GO:0006617 (P) SRP-dependent cotransla... GO:0048501 (C) signal recognition part... |
URS000080E293 | RF00169 | Escherichia coli 562 |
17086193 | Assembly1 protein:9:2~431 39,47~49,62~63 |
|
| 233 |
2j37:A (8.7Å) |
128 | GO:0006617 (P) SRP-dependent cotransla... GO:0005786 (C) signal recognition part... |
URS000080DE1E | RF00017 RF01856 |
Canis sp. 9616 |
17086193 | Assembly1 protein:B:14~120 141~143,147~151,195~198,203~20 4 protein:W:8~488 184~185,192~195,207~208 |
|
| 234 |
2jtp:A |
34 | GO:0075523 (P) viral translational fra... | URS000080E15C | RF01835 | synthetic construct 32630 |
17868691 | ||
| 235 |
2k4c:A |
76 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000009F4BF | RF00005 | Escherichia coli 562 |
18787959 | ||
| 236 |
2ke6:A |
48 | GO:0008298 (P) intracellular mRNA loca... GO:0051028 (P) mRNA transport |
URS000080DEE0 | RF00207 | Drosophila melanogaster 7227 |
20473315 | ||
| 237 |
2krl:A |
102 | GO:1902679 (P) negative regulation of ... | URS000080E1F0 | RF00500 | Turnip crinkle virus 11988 |
20080629 | ||
| 238 |
2kur:A |
48 | GO:0008298 (P) intracellular mRNA loca... GO:0051028 (P) mRNA transport |
URS000080E344 | RF00207 | Drosophila melanogaster 7227 |
20473315 | ||
| 239 |
2kuu:A |
48 | GO:0008298 (P) intracellular mRNA loca... GO:0051028 (P) mRNA transport |
URS000080E155 | RF00207 | Drosophila melanogaster 7227 |
20473315 | ||
| 240 |
2kuv:A |
48 | GO:0008298 (P) intracellular mRNA loca... GO:0051028 (P) mRNA transport |
URS000080DE75 | RF00207 | Drosophila melanogaster 7227 |
20473315 | ||
| 241 |
2kuw:A |
48 | GO:0008298 (P) intracellular mRNA loca... GO:0051028 (P) mRNA transport |
URS000080E2A2 | RF00207 | Drosophila melanogaster 7227 |
20473315 | ||
| 242 |
2l1f:A |
65 | GO:0019072 (P) viral genome packaging | URS000080DD56 | RF00374 | Moloney murine leukemia virus 11801 |
20933521 | Assembly1 rna:B:709~774 310,328,330~334,352~353,362,36 4~369 |
|
| 243 |
2l1f:B |
66 | GO:0019072 (P) viral genome packaging | URS000080DE60 | RF00374 | Moloney murine leukemia virus 11801 |
20933521 | Assembly1 rna:A:309~373 710,728,730~734,752~753,762,76 4~769 |
|
| 244 |
2l3j:B |
71 | GO:0035195 (P) miRNA-mediated post-tra... GO:0016442 (C) RISC complex |
URS000080DF7D | RF02095 | Rattus norvegicus 10116 |
20946981 | Assembly1 protein:A:1~236 245~248,255,258,260~261,268~26 9,277~279,287~288,290~293,299~ 300 |
|
| 245 |
2l94:A |
45 | GO:0075523 (P) viral translational fra... | URS000080E22A | RF00480 | Human immunodeficiency virus 1 11676 |
21648432 | Assembly1 L94:A:46 9~11,26~27,33~34 |
|
| 246 |
2lc8:A |
56 | GO:0039705 (P) viral translational rea... | URS0000A7636D | RF01073 | Moloney murine leukemia virus 11801 |
22121021 | ||
| 247 |
2lkr:A |
111 | GO:0030621 (F) U4 snRNA binding GO:0045131 (F) pre-mRNA branch point b... GO:0000244 (P) spliceosomal tri-snRNP ... GO:0000353 (P) formation of quadruple ... GO:0000348 (P) mRNA branch site recogn... GO:0005688 (C) U6 snRNP GO:0046540 (C) U4/U6 x U5 tri-snRNP co... GO:0005686 (C) U2 snRNP |
URS000080E147 | RF00026 RF00004 |
Saccharomyces cerevisiae 4932 |
22328579 | 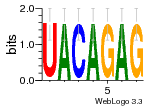 383 |
|
| 248 |
2lps:A |
34 | GO:0000373 (P) Group II intron splicing | URS000080DF0B | RF00029 | Saccharomyces cerevisiae 4932 |
22714631 14745440 |
||
| 249 |
2lpt:A |
34 | GO:0000373 (P) Group II intron splicing | URS000080E292 | RF00029 | synthetic construct 32630 |
22714631 16428604 |
||
| 250 |
2mf0:G |
72 | GO:0006109 (P) regulation of carbohydr... GO:0006401 (P) RNA catabolic process |
URS000080DEDE | RF00166 | Pseudomonas fluorescens 294 |
24828038 | Assembly1 protein:A:1~59 23,25,27~31,45,48~50,53 protein:B:1~59 21~23,26~28,30~32,45~53 protein:C:1~59 3~4,6,8~12,59~61,63~65,67~69 protein:D:1~59 2~4,6~9,12,60~61,63~68 protein:E:1~59 38~40,42~43 protein:F:1~59 38~42 |
|
| 251 |
2mf1:G |
72 | GO:0006109 (P) regulation of carbohydr... GO:0006401 (P) RNA catabolic process |
URS000080DEDE | RF00166 | Pseudomonas fluorescens 294 |
24828038 | Assembly1 protein:A:1~59 18~20,25,27~31,44~46,48~50,52~ 54 protein:B:1~59 21~23,26~32,46~53 protein:C:1~59 3~4,6,8~12,59~61,63~65,67~69 protein:D:1~59 2~9,12,60~61,63~68 protein:E:1~59 38~40,42~43,57~58,72 protein:F:1~59 38~43,72 |
|
| 252 |
2miy:A |
59 | GO:0008616 (P) queuosine biosynthetic ... | URS000080E2D6 | RF01054 | Streptococcus pneumoniae 1313 |
24469808 | Assembly1 PRF:A:101 7~8,19~20,50~51 |
|
| 253 |
2n1q:A |
155 | GO:0019074 (P) viral RNA genome packaging GO:0043484 (P) regulation of RNA splicing |
URS000086851D | RF01381 RF00175 RF01380 |
Human immunodeficiency virus 1 11676 |
25999508 | ||
| 254 |
2n7m:X |
92 | GO:0017070 (F) U6 snRNA binding GO:0000244 (P) spliceosomal tri-snRNP ... GO:0000353 (P) formation of quadruple ... GO:0005687 (C) U4 snRNP GO:0046540 (C) U4/U6 x U5 tri-snRNP co... |
URS00008FED30 | RF00015 | Saccharomyces cerevisiae 4932 |
26655855 | ||
| 255 |
2nqp:F (3.5Å) |
71 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS00004EADCF | RF00005 | Escherichia coli 562 |
17466622 | Assembly1 protein:C:7~270 16,19~20,56 protein:D:8~270 16,22~25,40 K:F:77 60~62 K:F:78 48~49,59~60 K:F:79 54~55,58 K:F:80 20,20A K:F:81 20,20A,48,59~60 |
|
| 256 |
2nr0:E (3.9Å) |
81 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS00004630EA | RF00005 | Escherichia coli 562 |
17466622 | Assembly1 protein:A:7~270 16,23,25,31,39~40 protein:B:7~270 19,56 |
|
| 257 |
2nr0:F (3.9Å) |
72 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS00004630EA | RF00005 | Escherichia coli 562 |
17466622 | Assembly1 protein:A:7~270 19~20,56 protein:B:7~270 16~17,23~25,33~34,36~41 |
|
| 258 |
2nr0:G (3.9Å) |
67 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS00004630EA | RF00005 | Escherichia coli 562 |
17466622 | Assembly2 protein:C:7~270 16~17,23~25,30,32,35~36,39~42 protein:D:7~270 16,19~20,56 |
|
| 259 |
2nr0:H (3.9Å) |
65 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS00004630EA | RF00005 | Escherichia coli 562 |
17466622 | Assembly2 protein:C:7~270 16,19~20,56 protein:D:7~270 16,23~25,31,39~40 |
|
| 260 |
2nre:F (4.0Å) |
56 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS00004630EA | RF00005 | Escherichia coli 562 |
17466622 | Assembly1 protein:A:7~270 16,23,25 K:F:78 17~18 |
|
| 261 |
2nz4:P (2.498Å) |
141 | GO:0003824 (F) catalytic activity GO:0010468 (P) regulation of gene expr... |
URS000080E011 | RF00234 | Bacillus anthracis 1392 |
17196404 | Assembly1 rna:E:-1~11 12,23~30,32~33,50,52~59,111~11 2 protein:A:7~96 14,16,17B,17C,17D,17E,17F,17G, 17H,17K,17L GLP:P:5001 42~43,57 MG:P:9010 28~30 |
|
| 262 |
2nz4:Q (2.498Å) |
141 | GO:0003824 (F) catalytic activity GO:0010468 (P) regulation of gene expr... |
URS000080E011 | RF00234 | Bacillus anthracis 1392 |
17196404 | Assembly2 rna:F:-1~11 12,23~30,32~33,50,52~57,59,111 ~112 protein:B:5~98 13,17B,17C,17D,17E,17F,17G,17H ,17K,17L GLP:F:5002 42~43,57~58 MG:Q:9012 28~30 |
|
| 263 |
2nz4:R (2.498Å) |
141 | GO:0003824 (F) catalytic activity GO:0010468 (P) regulation of gene expr... |
URS000080E011 | RF00234 | Bacillus anthracis 1392 |
17196404 | Assembly3 rna:G:-1~11 12,23~30,32~33,50,52~59,111~11 2 protein:C:8~97 12,14,17B,17C,17D,17E,17F,17G, 17H,17K,17L GLP:G:5003 42~43,57 MG:R:9014 29~30 |
|
| 264 |
2nz4:S (2.498Å) |
141 | GO:0003824 (F) catalytic activity GO:0010468 (P) regulation of gene expr... |
URS000080E011 | RF00234 | Bacillus anthracis 1392 |
17196404 | Assembly4 rna:H:-1~11 12,23~30,32~33,50,52~59,111~11 2 protein:D:7~96 14,17B,17C,17D,17E,17F,17G,17H ,17K,17L GLP:H:5004 42~43,57 MG:H:9016 29~30 |
|
| 265 |
2ob7:A (13.6Å) |
328 | GO:0006401 (P) RNA catabolic process | URS000080DFA2 | RF00023 | Thermus thermophilus 274 |
17179154 | ||
| 266 |
2om7:M (7.3Å) |
74 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000080E2DD | RF00005 | Thermus thermophilus 274 |
17349960 | Assembly1 rna:J:2094~2195 19,56 protein:K:18~223 54,63~64 |
|
| 267 |
2pxb:B (2.5Å) |
49 | GO:0006617 (P) SRP-dependent cotransla... GO:0048501 (C) signal recognition part... |
URS000080E210 | RF00169 | synthetic construct 32630 |
17637337 | Assembly1 protein:A:1~82 139~141,148~151,163~164 NCO:B:201 137,170~171 NCO:B:202 144~145,163 NCO:B:203 148~151 NCO:B:206 146,161~162 NCO:B:207 131~132,171 NCO:B:208 131~132,175 |
|
| 268 |
2pxd:B (2.0Å) |
49 | GO:0006617 (P) SRP-dependent cotransla... GO:0048501 (C) signal recognition part... |
URS000080E1B6 | RF00169 | synthetic construct 32630 |
17637337 | Assembly1 protein:A:1~82 140~141,148~151,163~164 NCO:B:201 137,170 NCO:B:202 144~146,163 NCO:B:203 148~151 NCO:B:206 146~147,162 NCO:B:207 131~133,175 |
|
| 269 |
2pxe:B (2.0Å) |
49 | GO:0006617 (P) SRP-dependent cotransla... GO:0048501 (C) signal recognition part... |
URS000080E2BA | RF00169 | synthetic construct 32630 |
17637337 | Assembly1 protein:A:1~82 139~141,148~151,163~164 NCO:B:201 137,170~171 NCO:B:202 144~146,163 NCO:B:203 148~151 NCO:B:206 147,161~162 NCO:B:207 131,133 |
|
| 270 |
2pxp:B (2.5Å) |
49 | GO:0006617 (P) SRP-dependent cotransla... GO:0048501 (C) signal recognition part... |
URS000080DFAC | RF00169 | synthetic construct 32630 |
17637337 | Assembly1 protein:A:1~82 139~141,148~151,163~164 NCO:B:201 170~171 NCO:B:202 144~146,163 NCO:B:203 148~151 NCO:B:206 146,161~162 NCO:B:207 131~132 |
|
| 271 |
2pxq:B (2.5Å) |
49 | GO:0006617 (P) SRP-dependent cotransla... GO:0048501 (C) signal recognition part... |
URS000080E073 | RF00169 | synthetic construct 32630 |
17637337 | Assembly1 protein:A:1~82 139~141,148~151,163~164 NCO:B:201 137,170~171 NCO:B:202 144~146,163 NCO:B:203 148~151 NCO:B:206 146~147,161~162 NCO:B:207 131,175~176 |
|
| 272 |
2pxt:B (2.5Å) |
49 | GO:0006617 (P) SRP-dependent cotransla... GO:0048501 (C) signal recognition part... |
URS000080E32F | RF00169 | synthetic construct 32630 |
17637337 | Assembly1 protein:A:1~82 139~141,148~151,163~164 NCO:B:201 137,170~171 NCO:B:202 144~145,163 NCO:B:203 148~150 NCO:B:206 146~147,162 NCO:B:207 175~176 |
|
| 273 |
2pxu:B (2.5Å) |
49 | GO:0006617 (P) SRP-dependent cotransla... GO:0048501 (C) signal recognition part... |
URS000080E2C9 | RF00169 | synthetic construct 32630 |
17637337 | Assembly1 protein:A:1~82 139~141,148~151,163~164 NCO:B:201 137,170~171 NCO:B:202 144~145,163~164 NCO:B:203 148~151 NCO:B:206 146~147,162 NCO:B:208 131~132,170~171 |
|
| 274 |
2qbz:X (2.6Å) |
153 | GO:0010468 (P) regulation of gene expr... GO:0032026 (P) response to magnesium ion |
URS000080E29A | RF00380 | Bacillus subtilis 1423 |
17803910 | Assembly1 MG:X:201 100,102~104 MG:X:202 23,100 MG:X:203 24~25 MG:X:206 80~81 K:X:301 27~29,155,157~158 K:X:302 66~67,74~75,106~107 K:X:303 25,72~73,161~162 K:X:304 23~24 |
|
| 275 |
2r8s:R (1.95Å) |
159 | GO:0000372 (P) Group I intron splicing | URS000080E2DA | RF00028 | synthetic construct 32630 |
18162543 | Assembly1 protein:L:1~214 130,167,169~173,195~197 protein:H:1~224 130~133,166~167,175~176,179 MG:R:1001 183~184,186 MG:R:1002 184,186~188 MG:R:1004 257~258 |
|
| 276 |
2rd2:B (2.6Å) |
74 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS0000115891 | RF00005 | Escherichia coli 562 |
18477696 | Assembly1 protein:A:8 902~903,906~907,911~913,916,92 5,927,934~938,971~976 |
|
| 277 |
2re8:B (2.6Å) |
74 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS0000115891 | RF00005 | Escherichia coli 562 |
18477696 | Assembly1 protein:A:8 902~904,906~907,910~914,925,92 7,934~938,972~976 |
|
| 278 |
2rkj:C (4.5Å) |
238 | GO:0000372 (P) Group I intron splicing | URS000080E20D | RF00028 | Twortvirus twort 55510 |
18172503 | Assembly1 rna:D:1~4 9~13,34,36~38,84,182~185,187 protein:B:39~416 47~51,88~91,109,123~124,203,21 2 |
|
| 279 |
2rkj:G (4.5Å) |
238 | GO:0000372 (P) Group I intron splicing | URS000080E20D | RF00028 | Twortvirus twort 55510 |
18172503 | Assembly2 rna:H:1~4 9~13,34,36~38,84,182~185,187 protein:F:39~416 47~48,50~52,88~91,109,123~124, 203,212 |
|
| 280 |
2rkj:K (4.5Å) |
238 | GO:0000372 (P) Group I intron splicing | URS000080E20D | RF00028 | Twortvirus twort 55510 |
18172503 | Assembly3 rna:L:1~4 9~13,34,36~38,84,182~185,187 protein:J:39~416 47~48,50~52,88~91,109,123~124, 203,212 |
|
| 281 |
2rkj:O (4.5Å) |
238 | GO:0000372 (P) Group I intron splicing | URS000080E20D | RF00028 | Twortvirus twort 55510 |
18172503 | Assembly4 rna:P:1~4 9~13,34,36~38,84,182~185,187 protein:N:39~416 47~48,50~51,88~91,109,123~124, 203,212 |
|
| 282 |
2tra:A (3.0Å) |
73 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS00005AD0B9 | RF00005 | Saccharomyces cerevisiae 4932 |
3272146 | ||
| 283 |
2v0g:B (3.5Å) |
76 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000080E15F | RF00005 | Thermus thermophilus 274 |
17588934 | Assembly1 protein:A:1~876 4~5,12~13,19~20,22~26,56,70,74 ~76 |
|
| 284 |
2v0g:F (3.5Å) |
76 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000080E15F | RF00005 | Thermus thermophilus 274 |
17588934 | Assembly2 protein:D:1~876 4,12~13,19~20,20A,22~25,39,56, 72,74~76 |
|
| 285 |
2v3c:M (2.5Å) |
96 | GO:0006617 (P) SRP-dependent cotransla... GO:0048501 (C) signal recognition part... |
URS000080DDBC | RF00169 | Methanocaldococcus jannaschii 2190 |
17846429 | Assembly1 protein:A:1~87 157~167,206~208,215~216 protein:C:2~427 142,187,195,203~206,213,218~22 2,232~233 |
|
| 286 |
2v3c:N (2.5Å) |
96 | GO:0006617 (P) SRP-dependent cotransla... GO:0048501 (C) signal recognition part... |
URS000080DDBC | RF00169 | Methanocaldococcus jannaschii 2190 |
17846429 | Assembly2 protein:B:1~87 157~160,162~167,206~209,216 protein:D:2~427 142,195~196,203~205,218~220 |
|
| 287 |
2vaz:A (10.0Å) |
82 | GO:0045974 (P) regulation of translati... | URS0002335951 | RF00114 | Escherichia coli 562 |
17889647 | ||
| 288 |
2xkv:B (13.5Å) |
96 | GO:0006617 (P) SRP-dependent cotransla... GO:0048501 (C) signal recognition part... |
URS000080E14C | RF00169 | Escherichia coli 562 |
21151118 | Assembly1 protein:C:23~65 38~40,47~50,63~64 |
|
| 289 |
2xnw:A (1.5Å) |
65 | GO:0009113 (P) purine nucleobase biosy... | URS000080DFC3 | RF00167 | Bacillus subtilis 1423 |
21439477 | Assembly1 NCO:A:1081 66~67 NCO:A:1082 39,56,69 NCO:A:1083 30~32,66 NCO:A:1084 46~47 NCO:A:1085 72~73 NCO:A:1086 37~38 NCO:A:1087 57~58 ACT:A:1088 32~33,38~39,66 ZZR:A:1089 21~22,47,51~52,74~75 |
|
| 290 |
2xnz:A (1.59Å) |
65 | GO:0009113 (P) purine nucleobase biosy... | URS000080DFC3 | RF00167 | Bacillus subtilis 1423 |
21439477 | Assembly1 NCO:A:1082 39,56,69 NCO:A:1083 30~32,66 NCO:A:1085 57~58 NCO:A:1086 24,71 ACT:A:1087 32~33,38~39,66 3AW:A:1088 21~22,47,51~52,74~75 K:A:1090 20,48~49 K:A:1091 37~38 K:A:1094 26,69 |
|
| 291 |
2xo0:A (1.7Å) |
65 | GO:0009113 (P) purine nucleobase biosy... | URS000080DFC3 | RF00167 | Bacillus subtilis 1423 |
21439477 | Assembly1 ACT:A:102 32~33,38~39,66 NCO:A:103 28,66~67 NCO:A:104 39,56,69 NCO:A:105 30~32,66 NCO:A:106 37~38 NCO:A:107 24,71 K:A:108 41,54 K:A:109 51~52,74~75 K:A:113 20,49 ZZS:A:110 21~22,51~52,74~75 |
|
| 292 |
2xo1:A (1.6Å) |
65 | GO:0009113 (P) purine nucleobase biosy... | URS00008FED1E | RF00167 | Bacillus subtilis 1423 |
21439477 | Assembly1 N6M:A:1081 21~22,47,51~52,74~75 NCO:A:1083 39,56,69 NCO:A:1084 30~32,66 NCO:A:1085 57~58 NCO:A:1087 24,71 NCO:A:1088 37~38 ACT:A:1089 32~33,38~39 K:A:1091 41,53 K:A:1092 26,69 |
|
| 293 |
2xxa:F (3.94Å) |
102 | GO:0006617 (P) SRP-dependent cotransla... GO:0048501 (C) signal recognition part... |
URS00003F3490 | RF00169 | Deinococcus radiodurans 1299 |
21330537 | Assembly2 protein:C:7~431 34,42~45,57~59,83 protein:D:20~302 10,83,85,94~97 |
|
| 294 |
2xxa:G (3.94Å) |
102 | GO:0006617 (P) SRP-dependent cotransla... GO:0048501 (C) signal recognition part... |
URS00003F3490 | RF00169 | Deinococcus radiodurans 1299 |
21330537 | Assembly1 protein:A:4~431 34~35,42~45,57~59,83 protein:B:20~302 10,83,94~97 |
|
| 295 |
2ydh:A (2.9Å) |
94 | GO:0010468 (P) regulation of gene expr... | URS000080E1B3 | RF00162 | Caldanaerobacter subterraneus subsp. tengcongensis 119072 |
21893284 | Assembly1 rna:A-2:1~94 32~34 BA:A:1095 26~28 BA:A:1097 10,64 SAM:A:1100 7~8,11,45,47,57~59,88~89 |
|
| 296 |
2ygh:A (2.6Å) |
95 | GO:0010468 (P) regulation of gene expr... | URS0000BC4670 | RF00162 | Caldanaerobacter subterraneus subsp. tengcongensis 119072 |
21893284 | Assembly1 SAM:A:1096 7~8,11,45~47,57~59,88~89 NA:A:1097 10~11,64 |
|
| 297 |
2z74:B (2.2Å) |
123 | GO:0003824 (F) catalytic activity GO:0010468 (P) regulation of gene expr... |
URS000080DF18 | RF00234 | Caldanaerobacter subterraneus subsp. tengcongensis 119072 |
17804015 | Assembly1 rna:A:-1~20 23~35,37,39~40,58,60~67,115~11 7 G6P:A:201 65~66 MG:B:301 35~36 MG:B:302 57,59 |
|
| 298 |
2z75:B (1.7Å) |
123 | GO:0003824 (F) catalytic activity GO:0010468 (P) regulation of gene expr... |
URS000080DF18 | RF00234 | Caldanaerobacter subterraneus subsp. tengcongensis MB4 273068 |
17804015 | Assembly1 rna:A:-1~20 23~37,39~40,58,60~65,67,115~11 7 GLP:A:201 50,65~66 MG:B:301 36~37 MG:B:302 79,81 MG:B:303 57,59 MG:B:307 60~61 MG:B:308 60~61 |
|
| 299 |
2z9q:A (11.7Å) |
75 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000080E1EE | RF00005 | Thermus aquaticus 271 |
12859903 | ||
| 300 |
2zm5:C (2.55Å) |
74 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS00005AA258 | RF00005 | Escherichia coli K-12 83333 |
19435325 | Assembly1 protein:A:9~314 26~41 MG:C:77 17,19 MG:C:79 7~8,14 MG:C:81 58~59 MG:C:82 22,46 |

Download all results in tab-seperated text for
3576 RNAs.
Click the entry in the '#' column to view structure details of that RNA chain.
Hover over PDB to view the title of the structure.
Hover over Rfam to view names of Rfam families.
Hover over PubMed to view title of the PubMed publications.
Hover over Ligand to view the name of the ligand and the ligand-binding nucleotides on the RNA.
<<
<
1 2 3 4 5 6 7 8 9 10 11 12 13
>
>>
<<
<
1 2 3 4 5 6 7 8 9 10 11 12 13
>
>>
Reference:
