| # | PDB (Resolution) |
Length | EC number & GO term |
RNAcentral | Rfam | Taxon | PubMed | ATtRACT motif |
Ligand & binding nucleotides |
|---|---|---|---|---|---|---|---|---|---|
| 301 |
2zm5:D (2.55Å) |
69 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS00005AA258 | RF00005 | Escherichia coli K-12 83333 |
19435325 | Assembly2 protein:B:7~311 26~30,32~41 |
|
| 302 |
2zue:B (2.0Å) |
76 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS00000D2896 | RF00005 | Pyrococcus horikoshii 53953 |
19656186 | Assembly1 protein:A:2~629 905,907,913~914,919~920,920A,9 23~925,935~941,956,969~972,974 |
|
| 303 |
2zuf:B (2.3Å) |
76 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS00000D2896 | RF00005 | synthetic construct 32630 |
19656186 | Assembly1 protein:A:2~629 904~905,907,913~914,919~920,92 0A,923~925,935~941,956,970~972 ,974 |
|
| 304 |
2zxu:C (2.75Å) |
74 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS00005AA258 | RF00005 | Escherichia coli K-12 83333 |
19435325 | Assembly1 protein:A:9~314 26~30,32~41 MG:C:77 17,19 MG:C:79 7~8,14 MG:C:81 58~59 MG:C:82 22,46 |
|
| 305 |
2zxu:D (2.75Å) |
69 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS00005AA258 | RF00005 | Escherichia coli K-12 83333 |
19435325 | Assembly2 protein:B:7~311 26~41 |
|
| 306 |
2zzm:B (2.65Å) |
84 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS00001612D2 | RF00005 | Methanocaldococcus jannaschii 2190 |
19749755 | Assembly1 protein:A:2~334 11~12,19~20,22~26,34~41,56~57 |
|
| 307 |
2zzn:C (2.95Å) |
71 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS00000B8EA0 | RF00005 | Methanocaldococcus jannaschii 2190 |
19749755 | Assembly1 protein:A:1~336 11~13,19~23,25~26,32~34,36~41, 56~57,69 |
|
| 308 |
2zzn:D (2.95Å) |
71 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS00000B8EA0 | RF00005 | Methanocaldococcus jannaschii 2190 |
19749755 | Assembly2 protein:B:2~336 11~12,18~20,25~26,32~34,36~41, 56~57 |
|
| 309 |
3a2k:C (3.65Å) |
77 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000080E29F | RF00005 | Bacillus subtilis 1423 |
19847269 | Assembly1 protein:A:1~462 1,10~13,26~27,33~40,67~73,75 |
|
| 310 |
3a2k:D (3.65Å) |
77 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000080E29F | RF00005 | Bacillus subtilis 1423 |
19847269 | Assembly2 protein:B:1~462 1,10~11,26~27,33~40,67~70,72~7 5 |
|
| 311 |
3a3a:A (3.1Å) |
86 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000014E598 | RF01852 RF00005 |
Homo sapiens 9606 |
19692584 |  1285  s70  787  757 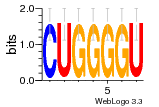 805 |
|
| 312 |
3adb:C (2.8Å) |
92 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000080DE1A | RF01852 | Methanopyrus kandleri 2320 |
20705242 | Assembly1 protein:A:-1~248 1,13,15,19~20,20A,21~24,41,47M ,56,68~69,75~76 |
|
| 313 |
3adb:D (2.8Å) |
92 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000080DE1A | RF01852 | Methanopyrus kandleri 2320 |
20705242 | Assembly1 protein:B:-10~248 1~2,15,19~20,20A,21~22,42,56,6 8~71,73~76 |
|
| 314 |
3adc:C (2.9Å) |
88 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000080DE1A | RF01852 | Methanopyrus kandleri 2320 |
20705242 | Assembly1 protein:A:-1~248 1~2,15,19~20,20A,21~23,41,68~7 1,73~76 |
|
| 315 |
3adc:D (2.9Å) |
92 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000080DE1A | RF01852 | Methanopyrus kandleri 2320 |
20705242 | Assembly1 protein:B:-1~248 1~2,16,19~20,20A,22,56,69~76 |
|
| 316 |
3add:C (2.4Å) |
88 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000080DE1A | RF01852 | Methanopyrus kandleri 2320 |
20705242 | Assembly1 protein:A:-2~248 1,14~15,19~20,20A,21~23,41,47M ,68~69,75~76 |
|
| 317 |
3add:D (2.4Å) |
88 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000080DE1A | RF01852 | Methanopyrus kandleri 2320 |
20705242 | Assembly1 protein:B:-1~248 1~2,15,19~20,20A,56,68~71,73~7 4 |
|
| 318 |
3akz:E (2.9Å) |
74 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000002EA11 | RF00005 | Thermotoga maritima 2336 |
20882017 | Assembly4 protein:A:24~486 3~7,11~13,24~25,34~39,68~71,74 ~76 |
|
| 319 |
3akz:F (2.9Å) |
74 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000002EA11 | RF00005 | Thermotoga maritima 2336 |
20882017 | Assembly1 protein:B:24~486 3~8,11~13,24~26,34~38,69~71,74 ~76 |
|
| 320 |
3akz:G (2.9Å) |
74 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000002EA11 | RF00005 | Thermotoga maritima 2336 |
20882017 | Assembly3 protein:C:24~486 3~6,11~13,24~26,34~39,69~71,74 ~76 |
|
| 321 |
3akz:H (2.9Å) |
74 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000002EA11 | RF00005 | Thermotoga maritima 2336 |
20882017 | Assembly2 protein:D:24~486 3~7,11~13,25~26,34~39,69~71,74 ~76 |
|
| 322 |
3al0:E (3.368Å) |
74 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000002EA11 | RF00005 | synthetic construct 32630 |
20882017 | Assembly1 protein:B:1~482 1,18~20,54,56,61~63 protein:C:3~590 4~6,12~14,24~25,34~38,69~72,74 ~76 |
|
| 323 |
3amt:B (2.9Å) |
78 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000080E317 | RF00005 | Archaeoglobus fulgidus 2234 |
22002223 | Assembly1 protein:A:0~420 1,24~26,33~40,69~72 |
|
| 324 |
3amu:B (3.1Å) |
78 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000080E317 | RF00005 | Archaeoglobus fulgidus 2234 |
22002223 | Assembly1 protein:A:0~420 24~27,31,33~34,36~40,69~72 |
|
| 325 |
3b0v:A (3.51Å) |
73 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS0000490B64 | RF00005 | Thermus thermophilus HB8 300852 |
22123979 | Assembly1 protein:C:1 14,16,19~20,22~25,39~42,56~57 |
|
| 326 |
3b0v:B (3.51Å) |
73 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS0000490B64 | RF00005 | Thermus thermophilus HB8 300852 |
22123979 | Assembly2 protein:D:1 14,17,19~20,22~25,39~42,56~57 |
|
| 327 |
3b4a:B (2.7Å) |
122 | GO:0003824 (F) catalytic activity GO:0010468 (P) regulation of gene expr... |
URS000080E06F | RF00234 | Caldanaerobacter subterraneus subsp. tengcongensis 119072 |
17990888 | Assembly1 rna:A:-2~20 24~35,37,39~40,58,60~67,115~11 7 GLP:A:401 51,65 MG:B:201 36~37 MG:B:202 38~39 MG:B:203 56~57,59 |
|
| 328 |
3b4b:B (2.7Å) |
123 | GO:0003824 (F) catalytic activity GO:0010468 (P) regulation of gene expr... |
URS000080E06F | RF00234 | Caldanaerobacter subterraneus subsp. tengcongensis 119072 |
17990888 | Assembly1 rna:A:1~20 23~37,39~40,58,60~65,115~117 GLP:A:201 65~66 3AD:A:-1 40,65~67 MG:B:301 36~37 |
|
| 329 |
3b4c:B (3.0Å) |
121 | GO:0003824 (F) catalytic activity GO:0010468 (P) regulation of gene expr... |
URS000080DF18 | RF00234 | Caldanaerobacter subterraneus subsp. tengcongensis 119072 |
17990888 | Assembly1 rna:A:1~20 23~37,39~40,58,60~65,115~117 3AD:A:-1 40,65~67 MG:B:301 36~37 |
|
| 330 |
3bbv:z (10.0Å) |
76 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS00005AA258 | RF00005 | Thermus thermophilus 274 |
19013177 | ||
| 331 |
3bo2:B (3.31Å) |
197 | GO:0000372 (P) Group I intron splicing | URS000080E37C | RF00028 | synthetic construct 32630 |
18408159 | Assembly1 rna:C:191~206 61~62,83~85,127~128,130,172,17 9~190 rna:D:1~6 4~10,59~60,87 rna:E:1~3 10~12,37,87~88,168~169 protein:A:4~98 105,1003~1009,1012~1013 MG:B:1 88,170,172 MG:B:2 128,172 MG:B:3 173~174 MG:B:1016 124,171 MG:B:1018 38~39 MG:B:1019 48,133 |
|
| 332 |
3bo3:B (3.4Å) |
197 | GO:0000372 (P) Group I intron splicing | URS000080E37C | RF00028 | synthetic construct 32630 |
18408159 | Assembly1 rna:C:191~206 61~62,83~84,127~128,130,172~17 3,178,180~190 rna:D:1~9 4~12,37,59~60,87~88,167~169,17 2 protein:A:4~98 1003~1009,1012~1013 MG:B:1 88,170,172 MG:B:2 128,172 MG:B:3 173~174 MG:B:1020 124,171 MG:B:1021 38~39 K:B:1015 22~24 |
|
| 333 |
3bo4:B (3.33Å) |
197 | GO:0000372 (P) Group I intron splicing | URS000080E37C | RF00028 | synthetic construct 32630 |
18408159 | Assembly1 rna:C:191~206 61~62,83~84,127~128,130,172,17 9~190 rna:D:1~9 4~12,37~38,60,87,168 protein:A:4~98 1003~1009,1012~1013 MG:B:1 88,170,172 MG:B:2 87,128,172 MG:B:3 173~174 MG:B:1022 124,170~171 MG:B:1023 38~39 K:B:1015 149~152 K:B:1016 124~127,171 K:B:1017 22~24 |
|
| 334 |
3bwp:A (3.1Å) |
356 | GO:0000373 (P) Group II intron splicing | URS000080DD31 | RF02001 RF01998 |
Oceanobacillus iheyensis 182710 |
18388288 | Assembly1 MG:A:413 375,377 MG:A:414 358~359,377 K:A:420 3~5,376 K:A:421 66~68,120 |
|
| 335 |
3cw1:V (5.493Å) |
138 | GO:0030627 (F) pre-mRNA 5'-splice site... GO:0000395 (P) mRNA 5'-splice site rec... GO:0005685 (C) U1 snRNP |
URS000080E05C | RF00003 | Homo sapiens 9606 |
19325628 10025403 21516107 15312772 10931958 7984237 2374709 |
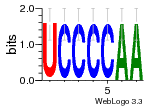 121 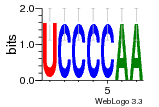 760 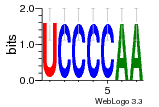 1031  s52  509 |
|
| 336 |
3cw1:v (5.493Å) |
138 | GO:0030627 (F) pre-mRNA 5'-splice site... GO:0000395 (P) mRNA 5'-splice site rec... GO:0005685 (C) U1 snRNP |
URS000080E05C | RF00003 | Homo sapiens 9606 |
19325628 10025403 21516107 15312772 10931958 7984237 2374709 |
 121  760  1031  s52  509 |
|
| 337 |
3cw1:w (5.493Å) |
138 | GO:0030627 (F) pre-mRNA 5'-splice site... GO:0000395 (P) mRNA 5'-splice site rec... GO:0005685 (C) U1 snRNP |
URS000080E05C | RF00003 | Homo sapiens 9606 |
19325628 10025403 21516107 15312772 10931958 7984237 2374709 |
 121  760  1031  s52  509 |
|
| 338 |
3cw1:x (5.493Å) |
138 | GO:0030627 (F) pre-mRNA 5'-splice site... GO:0000395 (P) mRNA 5'-splice site rec... GO:0005685 (C) U1 snRNP |
URS000080E05C | RF00003 | Homo sapiens 9606 |
19325628 10025403 21516107 15312772 10931958 7984237 2374709 |
 121  760  1031  s52  509 |
|
| 339 |
3cw5:A (3.1Å) |
77 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS00001BBAFC | RF00005 | Escherichia coli 562 |
18653533 | ||
| 340 |
3cw6:A (3.3Å) |
77 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS00001BBAFC | RF00005 | Escherichia coli 562 |
18653533 | ||
| 341 |
3d0u:A (2.8Å) |
161 | GO:0009085 (P) lysine biosynthetic pro... | URS000080DFA3 | RF00168 | Thermotoga maritima 2336 |
18593706 | Assembly1 IRI:A:202 138~139,148~149 IRI:A:203 120~122 IRI:A:204 80~81 LYS:A:205 8~9,77,111~112,152 |
|
| 342 |
3d0x:A (2.95Å) |
161 | GO:0009085 (P) lysine biosynthetic pro... | URS000080DFA3 | RF00168 | Thermotoga maritima 2336 |
18593706 | ||
| 343 |
3deg:A (10.9Å) |
76 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000011107D | RF00005 | Escherichia coli 562 |
19172743 | Assembly1 rna:J:1944~1961 75~76 rna:K:2547~2561 74,76 protein:C:1~545 8~12,66~71 |
|
| 344 |
3deg:B (10.9Å) |
77 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000041AF00 | RF00005 | Escherichia coli 562 |
19172743 | ||
| 345 |
3dhs:A (3.6Å) |
215 | EC:3.1.26.5 ribonuclease P GO:0004526 (F) ribonuclease P activity GO:0008033 (P) tRNA processing GO:0030680 (C) dimeric ribonuclease P ... |
URS000080DF78 | RF00011 | Geobacillus stearothermophilus 1422 |
19095619 | ||
| 346 |
3dig:X (2.8Å) |
174 | GO:0009085 (P) lysine biosynthetic pro... | URS000080E0C5 | RF00168 | Thermotoga maritima 2336 |
18784651 | Assembly1 K:X:200 11~12,14,115 NA:X:201 20,71 NA:X:203 25,67~69 NA:X:205 11~12 NA:X:212 123~125 SLZ:X:175 11~12,80~81,114~115,163 |
|
| 347 |
3dil:A (1.9Å) |
174 | GO:0009085 (P) lysine biosynthetic pro... | URS000080E0C5 | RF00168 | Thermotoga maritima 2336 |
18784651 | Assembly1 LYS:A:175 11~12,79~81,114~115,163 K:A:200 11~12,14,115 K:A:231 86,107~108 NA:A:201 20,71 NA:A:202 25,67~69 NA:A:204 11~12 NA:A:210 80,113,163~164 NA:A:215 172~173 NA:A:216 122~123 NA:A:220 121~122 NA:A:223 92~93 NA:A:225 69~70,130~131 1PE:A:250 142,159~160 1PE:A:251 141,160~161 |
|
| 348 |
3dim:A (2.9Å) |
174 | GO:0009085 (P) lysine biosynthetic pro... | URS000080E0C5 | RF00168 | Thermotoga maritima 2336 |
18784651 | Assembly1 CS:A:200 11~12,14,115 CS:A:250 141~142,160 NA:A:201 20,71 NA:A:203 124~125 NA:A:204 11~12 NA:A:207 146~147 NA:A:208 172~173 LYS:A:175 11~12,79~80,114~115,163 |
|
| 349 |
3dio:X (2.4Å) |
174 | GO:0009085 (P) lysine biosynthetic pro... | URS000080E0C5 | RF00168 | Thermotoga maritima 2336 |
18784651 | Assembly1 K:X:200 11~12,14,115 NA:X:201 20,71 NA:X:202 25,67~69 NA:X:203 11~12 NA:X:205 80,113,163~164 NA:X:207 92~93 NA:X:208 168~169 NA:X:212 68~69 NA:X:213 69~70,130~131 NA:X:214 15~16,82~83 IRI:X:250 1,172~173 IRI:X:251 109~110 IRI:X:252 123~125 IRI:X:253 55~57 IRI:X:254 83~84,109 IRI:X:255 13,15,117,131 IRI:X:257 149~150 IRI:X:258 113~115 IRI:X:259 124~125,127 IRI:X:260 157~158 IRI:X:261 26~27 IRI:X:262 40,52~53 IRI:X:263 91~92,98 IRI:X:264 8~9 IRI:X:265 140,142,159~160 LYS:X:175 11~12,80~81,114~115,163 1PE:X:280 7,170 |
|
| 350 |
3diq:A (2.7Å) |
174 | GO:0009085 (P) lysine biosynthetic pro... | URS000080E0C5 | RF00168 | Thermotoga maritima 2336 |
18784651 | Assembly1 K:A:200 11~12,14,115 NA:A:202 10~12 NA:A:206 20,71 NA:A:207 67~69 NA:A:209 146~147 HRG:A:175 11~12,80~81,114~115,163 |
|
| 351 |
3dir:A (2.9Å) |
174 | GO:0009085 (P) lysine biosynthetic pro... | URS000080E0C5 | RF00168 | Thermotoga maritima 2336 |
18784651 | Assembly1 K:A:200 11~12,14,115 NA:A:203 20,71 NA:A:204 25,67~69 NA:A:205 10~12 NA:A:206 145~147 NA:A:207 2,172~173 NA:A:211 152~154 IEL:A:175 11~12,80~81,114~115,163 |
|
| 352 |
3dis:A (3.1Å) |
174 | GO:0009085 (P) lysine biosynthetic pro... | URS000080E0C5 | RF00168 | Thermotoga maritima 2336 |
18784651 | Assembly1 NA:A:201 20,71 NA:A:202 67~69 NA:A:206 48~49 NA:A:207 84~85,109 |
|
| 353 |
3dix:A (2.9Å) |
174 | GO:0009085 (P) lysine biosynthetic pro... | URS000080E0C5 | RF00168 | Thermotoga maritima 2336 |
18784651 | Assembly1 K:A:200 11~12,14,115 NA:A:201 20,71 NA:A:202 145~147 NA:A:203 67~69 LYS:A:175 11~12,80~81,114~115,163 |
|
| 354 |
3diy:A (2.71Å) |
174 | GO:0009085 (P) lysine biosynthetic pro... | URS000080E0C5 | RF00168 | Thermotoga maritima 2336 |
18784651 | Assembly1 K:A:200 11~12,14,115 NA:A:201 20,71 NA:A:202 67~69 NA:A:203 72~73 NA:A:205 11~12 NA:A:207 80,113,163~164 LYS:A:175 11~12,80~81,114~115,163 |
|
| 355 |
3diz:A (2.85Å) |
174 | GO:0009085 (P) lysine biosynthetic pro... | URS000080E0C5 | RF00168 | Thermotoga maritima 2336 |
18784651 | Assembly1 K:A:200 11~12,14,115 NA:A:201 20,71 NA:A:202 67~69 NA:A:205 15~16 NA:A:211 10,142,166~167 NA:A:212 152~153 LYS:A:175 11~12,79~80,114~115,163 |
|
| 356 |
3dj0:A (2.5Å) |
174 | GO:0009085 (P) lysine biosynthetic pro... | URS000080E0C5 | RF00168 | Thermotoga maritima 2336 |
18784651 | Assembly1 K:A:200 11~12,14,115 NA:A:201 20,71 NA:A:202 25,67~69 NA:A:204 11~12 NA:A:210 80,113,164 NA:A:213 24~25 NA:A:215 121~122 NA:A:216 123~125 OLZ:A:175 11~12,80~81,114~115,163 |
|
| 357 |
3dj2:A (2.5Å) |
174 | GO:0009085 (P) lysine biosynthetic pro... | URS000080E0C5 | RF00168 | Thermotoga maritima 2336 |
18784651 | Assembly1 NA:A:201 145~147 NA:A:202 20,71 NA:A:204 11~12 LYS:A:175 11~12,80~81,114~115,163 TL:A:250 80,113,164 TL:A:251 115~116 TL:A:253 43~45 TL:A:254 82~83 TL:A:255 122~123 TL:A:256 141~142,160~161 TL:A:257 171~173 TL:A:258 67~69 TL:A:259 11~12,14 |
|
| 358 |
3ds7:A (1.85Å) |
67 | GO:0009113 (P) purine nucleobase biosy... | URS000080E322 | RF00167 | synthetic construct 32630 |
19007790 | Assembly1 ACT:A:96 32~33,38~39,66 NCO:A:101 66~67 NCO:A:102 38,56 NCO:A:103 39,56,69 NCO:A:104 30~32,66 GNG:A:120 21~22,50~52,74~76 |
|
| 359 |
3ds7:B (1.85Å) |
67 | GO:0009113 (P) purine nucleobase biosy... | URS000080E322 | RF00167 | synthetic construct 32630 |
19007790 | Assembly2 ACT:B:296 232~233,238~239 NCO:B:301 266~267 NCO:B:302 238,256 NCO:B:303 239,256,269 NCO:B:304 230~232,266 NCO:B:305 237~238 NCO:B:306 226~227,268 GNG:B:320 221~222,251~252,274~276 |
|
| 360 |
3e5c:A (2.25Å) |
53 | EC:2.5.1.6 methionine adenosyltransferase GO:0004478 (F) methionine adenosyltran... GO:0006446 (P) regulation of translati... |
URS000080DEC9 | RF01767 | synthetic construct 32630 |
18806797 | Assembly1 SR:A:208 34,36 SR:A:213 14~15 SAM:A:216 7,31,36~38,47~48 |
|
| 361 |
3e5e:A (2.9Å) |
53 | EC:2.5.1.6 methionine adenosyltransferase GO:0004478 (F) methionine adenosyltran... GO:0006446 (P) regulation of translati... |
URS000080DEC9 | RF01767 | synthetic construct 32630 |
18806797 | Assembly1 SAH:A:220 7,37~38,47~48 |
|
| 362 |
3e5f:A (2.7Å) |
53 | EC:2.5.1.6 methionine adenosyltransferase GO:0004478 (F) methionine adenosyltran... GO:0006446 (P) regulation of translati... |
URS000080DEC9 | RF01767 | synthetic construct 32630 |
18806797 | Assembly1 SR:A:208 34,36 EEM:A:216 7,30~31,36~38,47~48 |
|
| 363 |
3eog:A (3.391Å) |
388 | GO:0000373 (P) Group II intron splicing | URS000080DD31 | RF02001 RF01998 |
Oceanobacillus iheyensis 182710 |
18953333 | Assembly1 rna:B:1~6 106,181~184,223,359,374~375 MG:A:413 119,239 MG:A:414 375,377 MG:A:415 358~359,377 MG:A:417 110~111 MG:A:423 66~67 MG:A:424 116~117 MG:A:426 154~156 MG:A:437 358~359,377 MG:A:440 4,375~376 MG:A:443 67~68 K:A:444 79~80 K:A:445 86~88,96~97 K:A:446 97~98 K:A:449 273,275 K:A:450 364~365 K:A:451 3~5,376 K:A:453 333~336 |
|
| 364 |
3eoh:A (3.125Å) |
381 | GO:0000373 (P) Group II intron splicing | URS000080DD31 | RF02001 RF01998 |
Oceanobacillus iheyensis 182710 |
18953333 | Assembly1 rna:B:1~6 106,181~184,223,358~359,374 MG:A:413 119,239 MG:A:414 358~359,377 MG:A:416 110~111 MG:A:422 66~67 MG:A:423 116~117 MG:A:425 154~156 MG:A:430 135~136 MG:A:434 286~287 MG:A:435 355~356 MG:A:436 358~359,377 MG:A:439 4,107,375~376 MG:A:440 320~321 MG:A:442 67~68 MG:B:7 375,377 K:A:443 79~80 K:A:444 86~88,95~97 K:A:445 97~98 K:A:448 273,275 K:A:449 364~365 K:A:450 3~5,376 |
|
| 365 |
3ep2:Y (9.0Å) |
74 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000080DD6E | RF00005 | Escherichia coli K-12 83333 |
19020518 | ||
| 366 |
3eph:E (2.95Å) |
69 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000080DDE1 | RF00005 | Saccharomyces cerevisiae 4932 |
18852462 | Assembly1 protein:A:13~421 9,20,22,26~45 |
|
| 367 |
3eph:F (2.95Å) |
69 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000080DDE1 | RF00005 | Saccharomyces cerevisiae 4932 |
18852462 | Assembly1 protein:B:13~421 20,26~44 |
|
| 368 |
3epj:E (3.1Å) |
69 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000080DDE1 | RF00005 | Saccharomyces cerevisiae 4932 |
18852462 | Assembly1 protein:A:13~421 20,22,26~44,46 |
|
| 369 |
3epj:F (3.1Å) |
69 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000080DDE1 | RF00005 | Saccharomyces cerevisiae 4932 |
18852462 | Assembly1 protein:B:13~421 26~44 MG:F:74 36~37 |
|
| 370 |
3epk:E (3.2Å) |
69 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000080DDE1 | RF00005 | Saccharomyces cerevisiae 4932 |
18852462 | Assembly1 protein:A:13~421 9,20,22,26~44 |
|
| 371 |
3epk:F (3.2Å) |
69 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000080DDE1 | RF00005 | Saccharomyces cerevisiae 4932 |
18852462 | Assembly1 protein:B:13~421 9,20,26~44 |
|
| 372 |
3epl:E (3.6Å) |
69 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000080DDE1 | RF00005 | Saccharomyces cerevisiae 4932 |
18852462 | Assembly1 protein:A:13~421 20,26~46 |
|
| 373 |
3epl:F (3.6Å) |
69 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000080DDE1 | RF00005 | Saccharomyces cerevisiae 4932 |
18852462 | Assembly1 protein:B:13~421 26,28~45 |
|
| 374 |
3eq3:Y (9.0Å) |
74 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000080DD6E | RF00005 | Escherichia coli K-12 83333 |
19020518 | ||
| 375 |
3eq4:Y (12.0Å) |
85 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000080DE10 | RF00005 | Escherichia coli K-12 83333 |
19020518 | ||
| 376 |
3fo4:A (1.9Å) |
63 | GO:0009113 (P) purine nucleobase biosy... | URS000080E134 | RF00167 | Bacillus subtilis 1423 |
19523903 | Assembly1 6GU:A:91 21~22,47,51~52,74~75 ACT:A:96 32~33,38~39 NCO:A:101 66~67 NCO:A:102 38~39,56 NCO:A:103 39,56,69 NCO:A:104 30~32,66 NCO:A:105 37~38 NCO:A:107 41,53~54 NCO:A:108 47,50~51 NCO:A:109 24,71 NCO:A:111 32~33 |
|
| 377 |
3fo6:A (1.9Å) |
67 | GO:0009113 (P) purine nucleobase biosy... | URS000080DD54 | RF00167 | Bacillus subtilis 1423 |
19523903 | Assembly1 6GO:A:91 21~22,47,51~52,74~75 ACT:A:96 32~33,38~39 NCO:A:101 66~67 NCO:A:102 38~39,56 NCO:A:103 56,69 NCO:A:104 30~32,66 NCO:A:105 37~38 NCO:A:107 41,53~54 NCO:A:108 46~47 NCO:A:109 24,71 |
|
| 378 |
3foz:C (2.5Å) |
74 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000080DF41 | RF00005 | Escherichia coli 562 |
19158097 | Assembly1 protein:A:7~311 26~41 protein:B:9~311 70~72 CA:C:76 58~59 CA:C:77 17,19 CA:C:78 7~8,14 |
|
| 379 |
3foz:D (2.5Å) |
69 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000080E12B | RF00005 | Escherichia coli 562 |
19158097 | Assembly1 protein:B:9~311 25~41 CA:D:1 22,45 CA:D:75 17,19 CA:D:77 44~45 CA:D:80 71~72 CA:D:81 7~8,14 |
|
| 380 |
3fu2:A (2.85Å) |
32 | GO:0008616 (P) queuosine biosynthetic ... | URS000080E020 | RF00522 | synthetic construct 32630 |
19234468 | Assembly1 PRF:A:101 5,11~12,17~18,30 CA:A:901 7,9 |
|
| 381 |
3fu2:B (2.85Å) |
33 | GO:0008616 (P) queuosine biosynthetic ... | URS000080E020 | RF00522 | synthetic construct 32630 |
19234468 | Assembly2 PRF:B:101 5,11~12,17~18,30 CA:B:903 7,9 |
|
| 382 |
3fu2:C (2.85Å) |
32 | GO:0008616 (P) queuosine biosynthetic ... | URS000080E020 | RF00522 | synthetic construct 32630 |
19234468 | Assembly3 PRF:C:101 5,11~12,17~18,30 CA:C:904 7,9 |
|
| 383 |
3g4m:A (2.4Å) |
67 | GO:0009113 (P) purine nucleobase biosy... | URS000080DDA1 | RF00167 | Bacillus subtilis 1423 |
19523903 | Assembly1 2BP:A:91 21~22,47,51~52,74~75 ACT:A:96 32~33,38,66 NCO:A:101 66~67 NCO:A:102 38~39,56 NCO:A:103 39,55~56,69 NCO:A:104 30~32,66 NCO:A:105 37~38 NCO:A:107 53~54 NCO:A:110 21~22 NCO:A:112 72~73 |
|
| 384 |
3g78:A (2.8Å) |
389 | GO:0000373 (P) Group II intron splicing | URS000080DD31 | RF02001 RF01998 |
Oceanobacillus iheyensis 182710 |
20823550 18388288 18953333 |
Assembly1 rna:Z:1~9 106,181~186,223,359,374,377 MG:A:413 287,380 MG:A:414 3~4 MG:A:423 360,374 MG:A:435 119,239 MG:A:439 355~356 MG:A:443 66~67 MG:A:445 181,223 MG:A:452 275~276 MG:A:455 60,198 MG:Z:10 375,377 MG:Z:11 358~359,377 K:A:461 4,106~107,375~376 K:A:462 65~68 K:A:463 115~117 K:A:464 274~275 K:A:465 25,27~28 K:A:466 251~252 K:A:467 184~185 K:A:468 333~335 K:A:469 263~265 K:A:470 297,319~321 K:A:471 283~284 K:A:472 192~194 K:A:474 86~88,95~97 K:A:475 3~5,376 K:A:476 79~80,103 K:A:477 32~34 K:A:478 228~229 K:A:480 288,358~359,377 K:A:481 330~332 K:A:482 348~351 K:A:483 120~121 K:A:484 153~155 K:A:485 88~90 K:A:486 56,58 |
|
| 385 |
3g8s:P (3.1Å) |
141 | GO:0003824 (F) catalytic activity GO:0010468 (P) regulation of gene expr... |
URS000080E011 | RF00234 | Bacillus anthracis 1392 |
19228039 | Assembly1 protein:A:7~96 13,17B,17C,17D,17E,17F,17G,17H ,17L rna:E:-1~11 12,23~30,33,44~45,52~57,59,111 ~112 MG:P:2 29~30 |
|
| 386 |
3g8s:Q (3.1Å) |
141 | GO:0003824 (F) catalytic activity GO:0010468 (P) regulation of gene expr... |
URS000080E011 | RF00234 | Bacillus anthracis 1392 |
19228039 | Assembly2 protein:B:5~98 13~14,17B,17C,17D,17E,17F,17G, 17H,17K,17L,110 rna:F:-1~11 12,23~30,32~33,44~45,50,52~59, 111~112 MG:Q:3 28~30 |
|
| 387 |
3g8s:R (3.1Å) |
141 | GO:0003824 (F) catalytic activity GO:0010468 (P) regulation of gene expr... |
URS000080E011 | RF00234 | Bacillus anthracis 1392 |
19228039 | Assembly3 protein:C:7~96 12~13,17B,17C,17D,17E,17F,17G, 17H,17K,17L,110 rna:G:-1~11 12,23~30,32~33,50,52~59,111~11 2 MG:R:6 46~47 MG:R:7 29~30 |
|
| 388 |
3g8s:S (3.1Å) |
141 | GO:0003824 (F) catalytic activity GO:0010468 (P) regulation of gene expr... |
URS000080E011 | RF00234 | Bacillus anthracis 1392 |
19228039 | Assembly4 protein:D:7~96 13,17B,17C,17D,17E,17F,17G,17H ,17K,17L rna:H:-1~11 12,23~30,32~33,52~57,59,111~11 2 MG:S:9 29~30 |
|
| 389 |
3g8t:P (3.0Å) |
141 | GO:0003824 (F) catalytic activity GO:0010468 (P) regulation of gene expr... |
URS000080E372 | RF00234 | synthetic construct 32630 |
19228039 | Assembly1 protein:A:7~96 17B,17C,17D,17E,17F,17G,17L rna:E:-1~11 12,23~28,30,32~33,44,52~57,59, 111~112 GLP:E:5001 42~43,57~58 MG:P:6 18~19 |
|
| 390 |
3g8t:Q (3.0Å) |
141 | GO:0003824 (F) catalytic activity GO:0010468 (P) regulation of gene expr... |
URS000080E372 | RF00234 | synthetic construct 32630 |
19228039 | Assembly2 protein:B:7~96 13,17B,17C,17E,17F,17G,17L,110 rna:F:-1~11 12,23~30,32~33,44,50,52~57,111 ~112 GLP:Q:5002 42~43,57~58 MG:Q:10 47~48 |
|
| 391 |
3g8t:R (3.0Å) |
141 | GO:0003824 (F) catalytic activity GO:0010468 (P) regulation of gene expr... |
URS000080E372 | RF00234 | synthetic construct 32630 |
19228039 | Assembly3 protein:C:7~96 13~14,17B,17C,17D,17E,17F,17G, 17H,17L rna:G:-1~11 12,23~30,32~33,44,50,52~58,111 ~112 GLP:R:5003 43,57 |
|
| 392 |
3g8t:S (3.0Å) |
141 | GO:0003824 (F) catalytic activity GO:0010468 (P) regulation of gene expr... |
URS000080E372 | RF00234 | synthetic construct 32630 |
19228039 | Assembly4 protein:D:7~94 14,17B,17C,17D,17E,17F,17G,17H ,17K,17L rna:H:-1~11 12,23~30,32~33,44,50,52~57,111 ~112 GLP:H:5004 42~43,57 |
|
| 393 |
3g96:P (3.01Å) |
141 | GO:0003824 (F) catalytic activity GO:0010468 (P) regulation of gene expr... |
URS000080E011 | RF00234 | synthetic construct 32630 |
19228039 | Assembly1 protein:A:7~96 17B,17C,17D,17E,17F,17G,17H,17 L rna:E:-1~11 12,23~30,32~33,44,52~58,111~11 2 |
|
| 394 |
3g96:Q (3.01Å) |
141 | GO:0003824 (F) catalytic activity GO:0010468 (P) regulation of gene expr... |
URS000080E011 | RF00234 | synthetic construct 32630 |
19228039 | Assembly2 protein:B:7~96 13,17B,17C,17D,17E,17F,17G,17H ,17K,17L rna:F:-1~11 12,23~30,32~33,50,52~59,111~11 2 6MN:F:12 42~43,57~58 |
|
| 395 |
3g96:R (3.01Å) |
141 | GO:0003824 (F) catalytic activity GO:0010468 (P) regulation of gene expr... |
URS000080E011 | RF00234 | synthetic construct 32630 |
19228039 | Assembly3 protein:C:7~96 12,17B,17C,17D,17E,17F,17G,17H ,17K,17L rna:G:-1~11 12,23~30,32~33,50,52~59,111~11 2 |
|
| 396 |
3g96:S (3.01Å) |
141 | GO:0003824 (F) catalytic activity GO:0010468 (P) regulation of gene expr... |
URS000080E011 | RF00234 | synthetic construct 32630 |
19228039 | Assembly4 protein:D:7~96 17B,17C,17D,17E,17F,17G,17H,17 L rna:H:-1~11 12,23~30,32~33,44,50,52~57,59, 111~112 |
|
| 397 |
3g9c:P (2.9Å) |
140 | GO:0003824 (F) catalytic activity GO:0010468 (P) regulation of gene expr... |
URS000080E011 | RF00234 | synthetic construct 32630 |
19228039 | Assembly1 protein:A:7~96 17B,17C,17D,17E,17F,17G,17H,17 L rna:E:1~11 12,23~30,50,52~57,111~112 GLP:E:5001 42~43,57~58 MG:P:1 28~29 |
|
| 398 |
3g9c:Q (2.9Å) |
141 | GO:0003824 (F) catalytic activity GO:0010468 (P) regulation of gene expr... |
URS000080E011 | RF00234 | synthetic construct 32630 |
19228039 | Assembly2 protein:B:7~96 12~13,17B,17C,17D,17E,17F,17G, 17H,17K,17L,110 rna:F:1~11 12,23~30,50,52~57,111~112 MG:Q:2 29~30 |
|
| 399 |
3g9c:R (2.9Å) |
141 | GO:0003824 (F) catalytic activity GO:0010468 (P) regulation of gene expr... |
URS000080E011 | RF00234 | synthetic construct 32630 |
19228039 | Assembly3 protein:C:7~96 17B,17C,17D,17E,17F,17G,17H,17 K,17L rna:G:1~11 12,23~30,50,52~57,111~112 MG:R:3 28~29 |
|
| 400 |
3g9c:S (2.9Å) |
140 | GO:0003824 (F) catalytic activity GO:0010468 (P) regulation of gene expr... |
URS000080E011 | RF00234 | synthetic construct 32630 |
19228039 | Assembly4 protein:D:7~96 14,17B,17C,17E,17F,17G,17H,17L rna:H:1~11 12,23~30,50,52~57,111~112 |

Download all results in tab-seperated text for
3576 RNAs.
Click the entry in the '#' column to view structure details of that RNA chain.
Hover over PDB to view the title of the structure.
Hover over Rfam to view names of Rfam families.
Hover over PubMed to view title of the PubMed publications.
Hover over Ligand to view the name of the ligand and the ligand-binding nucleotides on the RNA.
<<
<
1 2 3 4 5 6 7 8 9 10 11 12 13 14
>
>>
<<
<
1 2 3 4 5 6 7 8 9 10 11 12 13 14
>
>>
Reference:
