| # | PDB (Resolution) |
Length | EC number & GO term |
RNAcentral | Rfam | Taxon | PubMed | ATtRACT motif |
Ligand & binding nucleotides |
|---|---|---|---|---|---|---|---|---|---|
| 1 |
7p2e:A (2.4Å) |
955 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS0001A24B4C | RF00177 | Homo sapiens 9606 |
36480258 | 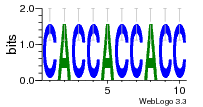 1321 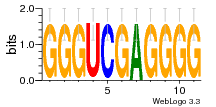 886 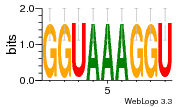 1302 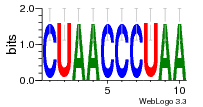 156  748  749  1207  M027_0.6  979  M089_0.6 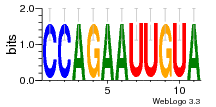 82 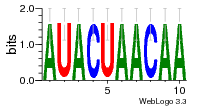 495 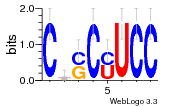 175 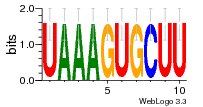 399 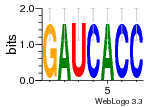 492 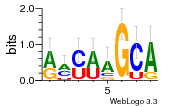 s44 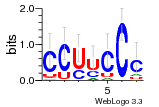 M043_0.6 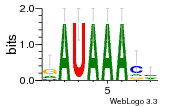 M033_0.6 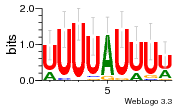 M329_0.6 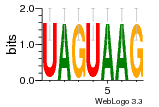 1194 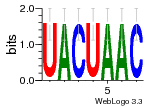 1197 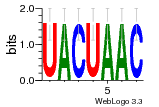 1196  938  M142_0.6  509  1290 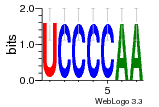 1031 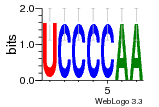 760 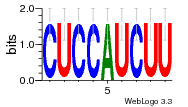 994 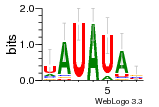 M164_0.6  1312  1273  779  778  s14  120  121  M047_0.6 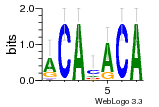 M169_0.6 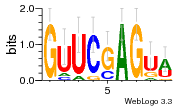 M274_0.6 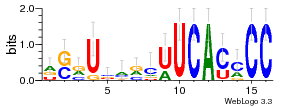 M325_0.6 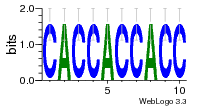 1354 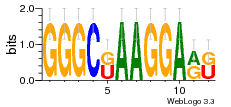 78 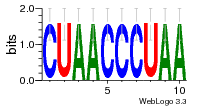 800 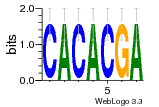 17  1186 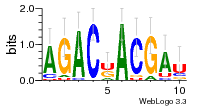 M331_0.6  1241  M143_0.6  1364  M016_0.6  33  s61 |
Assembly1 5I0:A:1703 655~656,898~899,1166~1167,1556 |
| 2 |
8cai:A (2.08Å) |
1114 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS0000D56C37 | RF00177 RF01959 RF02542 RF01960 |
Escherichia coli BW25113 679895 |
37550453 | Assembly1 5I0:A:1606 13~14,526~527,912~915,1490~149 1 |
|
| 3 |
8cgj:A (1.79Å) |
1130 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000268143B | RF00177 RF01959 RF02542 RF01960 |
Escherichia coli BW25113 679895 |
37550453 | Assembly1 5I0:A:1603 13~14,526~527,912~915,1490~149 1 |

Download all results in tab-seperated text for
3 RNAs.
Click the entry in the '#' column to view structure details of that RNA chain.
Hover over PDB to view the title of the structure.
Hover over Rfam to view names of Rfam families.
Hover over PubMed to view title of the PubMed publications.
Hover over Ligand to view the name of the ligand and the ligand-binding nucleotides on the RNA.
<<
<
1
>
>>
<<
<
1
>
>>
Reference:
