| # | PDB (Resolution) |
Length | EC number & GO term |
RNAcentral | Rfam | Taxon | PubMed | ATtRACT motif |
Ligand & binding nucleotides |
|---|---|---|---|---|---|---|---|---|---|
| 1 |
5aa0:AA (5.0Å) |
2889 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS0000868519 | RF02540 RF02541 RF00002 |
Thermus thermophilus HB8 300852 |
26283392 | Assembly1 8AN:AA:3002 2506~2507,2573,2583~2585 8AN:AA:3003 2251~2253 |
|
| 2 |
5aa0:BD (5.0Å) |
75 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000031D44F | RF00005 | Thermus thermophilus HB8 300852 |
26283392 | Assembly1 8AN:AA:3002 74~75 |
|
| 3 |
5tga:1 (3.3Å) |
3149 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS0000A7632E | RF02540 RF02543 |
Saccharomyces cerevisiae 4932 |
27827794 | 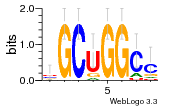 M107_0.6 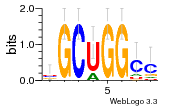 M080_0.6 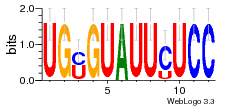 M299_0.6 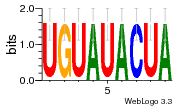 16 |
Assembly1 8AN:1:3403 2952~2954 |
| 4 |
5tga:5 (3.3Å) |
3150 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS0000A7632E | RF02540 RF02543 |
Saccharomyces cerevisiae 4932 |
27827794 |  M107_0.6  M080_0.6  M299_0.6  16 |
Assembly2 8AN:5:3403 2922,2952~2954 |
| 5 |
5tgm:1 (3.5Å) |
3149 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS0000B21DCE | RF02540 RF02543 |
Saccharomyces cerevisiae 4932 |
27827794 |  M107_0.6  M080_0.6  M299_0.6  16 |
Assembly1 8AN:P:101 2405,2619,2819~2820 |
| 6 |
5tgm:5 (3.5Å) |
3150 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS0000B21DEF | RF02540 RF02543 |
Saccharomyces cerevisiae 4932 |
27827794 |  M107_0.6  M080_0.6  M299_0.6  16 |
Assembly1 8AN:p:101 2405,2619,2819~2820 |
| 7 |
8eiu:a (2.24Å) |
2753 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS0002681390 | RF02541 | Escherichia coli 562 |
36660825 | Assembly1 8AN:Y:101 2506,2553,2583~2585,2602 8AN:Z:101 2251,2450~2451 |
|
| 8 |
8g6w:a (2.02Å) |
2753 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS00004B0F34 | RF02541 | Escherichia coli 562 |
37396857 | Assembly1 8AN:Z:101 2251,2450~2451 |
|
| 9 |
8g6x:a (2.31Å) |
2753 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS00004B0F34 | RF02541 | Escherichia coli 562 |
37396857 | Assembly1 8AN:Z:101 2251,2450~2451 |
|
| 10 |
8g6y:a (2.09Å) |
2753 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS00004B0F34 | RF02541 | Escherichia coli 562 |
37396857 | Assembly1 8AN:Z:101 2251,2450~2451 |

Download all results in tab-seperated text for
10 RNAs.
Click the entry in the '#' column to view structure details of that RNA chain.
Hover over PDB to view the title of the structure.
Hover over Rfam to view names of Rfam families.
Hover over PubMed to view title of the PubMed publications.
Hover over Ligand to view the name of the ligand and the ligand-binding nucleotides on the RNA.
<<
<
1
>
>>
<<
<
1
>
>>
Reference:
