| # | PDB (Resolution) |
Length | EC number & GO term |
RNAcentral | Rfam | Taxon | PubMed | ATtRACT motif |
Ligand & binding nucleotides |
|---|---|---|---|---|---|---|---|---|---|
| 1 |
3skz:A (2.605Å) |
68 | GO:0032546 (F) deoxyribonucleoside bin... GO:0031554 (P) regulation of terminati... |
URS000080DE91 | RF01510 | Mesoplasma florum 2151 |
21841796 | Assembly1 GMP:A:120 30~31,58~59,80~81 |
|
| 2 |
3skz:B (2.605Å) |
68 | GO:0032546 (F) deoxyribonucleoside bin... GO:0031554 (P) regulation of terminati... |
URS000080DE91 | RF01510 | Mesoplasma florum 2151 |
21841796 | Assembly2 GMP:B:120 30~31,54~55,58~59,80~81 |
|
| 3 |
4x4n:B (2.953Å) |
32 | N/A | URS000080E2D0 | N/A | Homo sapiens 9606 |
25640237 | 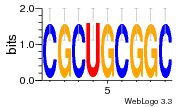 1124 |
Assembly1 GMP:B:101 15~16 |

Download all results in tab-seperated text for
3 RNAs.
Click the entry in the '#' column to view structure details of that RNA chain.
Hover over PDB to view the title of the structure.
Hover over Rfam to view names of Rfam families.
Hover over PubMed to view title of the PubMed publications.
Hover over Ligand to view the name of the ligand and the ligand-binding nucleotides on the RNA.
<<
<
1
>
>>
<<
<
1
>
>>
Reference:
