| # | PDB (Resolution) |
Length | EC number & GO term |
RNAcentral | Rfam | Taxon | PubMed | ATtRACT motif |
Ligand & binding nucleotides |
|---|---|---|---|---|---|---|---|---|---|
| 1 |
1y39:D (2.8Å) |
58 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000080E362 | RF02541 | Escherichia coli 562 |
15734647 | Assembly2 GOL:D:704 319,323,343~344,346~347 |
|
| 2 |
2qux:I (2.44Å) |
25 | N/A | URS000080DEBC | N/A | synthetic construct 32630 |
18066080 | Assembly3 GOL:I:26 11~12 |
|
| 3 |
2qux:L (2.44Å) |
25 | N/A | URS000080DEBC | N/A | synthetic construct 32630 |
18066080 | Assembly4 GOL:L:26 10~11 |
|
| 4 |
3dvz:A (1.0Å) |
27 | N/A | URS000080E28F | N/A | synthetic construct 32630 |
19228585 | Assembly1 GOL:A:1 2653~2654,2656 |
|
| 5 |
3dw4:A (0.97Å) |
27 | N/A | URS000080E28F | N/A | synthetic construct 32630 |
19228585 | Assembly1 GOL:A:1 2653~2654,2656 |
|
| 6 |
3dw6:A (1.0Å) |
27 | N/A | URS000080E28F | N/A | synthetic construct 32630 |
19228585 | Assembly1 GOL:A:1 2653~2654,2656 |
|
| 7 |
3rg5:A (2.0Å) |
86 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS0000624339 | RF01852 RF00005 |
Mus musculus 10090 |
21629646 | Assembly1 GOL:A:74 10,26 |
|
| 8 |
3rg5:B (2.0Å) |
86 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS0000624339 | RF01852 RF00005 |
Mus musculus 10090 |
21629646 | Assembly2 GOL:B:74 30~31 |
|
| 9 |
4al5:B (2.0Å) |
18 | N/A | URS000080DECD | N/A | Pseudomonas aeruginosa 287 |
22522703 | Assembly1 GOL:B:1022 19~20 |
|
| 10 |
4ed5:D (2.0Å) |
10 | N/A | URS000080E27D | N/A | synthetic construct 32630 |
23519412 | Assembly1 GOL:D:101 3~5 GOL:D:102 4,7 |
|
| 11 |
4nxh:A (1.158Å) |
27 | N/A | URS000080E119 | N/A | Escherichia coli 562 |
24766131 | Assembly1 GOL:A:2701 2653~2654,2656 |
|
| 12 |
5d99:A (0.97Å) |
27 | N/A | URS000080E28F | N/A | Enterobacteria phage L1 268588 |
27050127 19228585 |
Assembly1 GOL:A:2701 2653~2654,2656 |
|
| 13 |
5f8i:B (2.503Å) |
12 | N/A | URS000080DEB1 | N/A | synthetic construct 32630 |
27339134 | Assembly1 GOL:B:701 603~604 |
|
| 14 |
5f8m:B (2.83Å) |
12 | N/A | URS000080DEB1 | N/A | synthetic construct 32630 |
27339134 | Assembly1 GOL:B:701 603~604 |
|
| 15 |
5m73:A (3.4Å) |
145 | GO:0006617 (P) SRP-dependent cotransla... GO:0005786 (C) signal recognition part... |
URS0000A7784F | RF00017 RF01856 |
Homo sapiens 9606 |
27899666 |  1164  1212  M347_0.6  1373  s55  110  s71  s72 |
Assembly1 GOL:A:333 205~206 GOL:A:334 127,221~223 |
| 16 |
5on6:AR (3.1Å) |
3149 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000061F377 | RF02540 RF02543 |
Saccharomyces cerevisiae S288C 559292 |
29429877 | Assembly2 GOL:AR:4261 1529,1587,1590~1592 GOL:AR:4262 2793~2794 |
|
| 17 |
5tf6:B (2.3Å) |
71 | GO:0030621 (F) U4 snRNA binding GO:0000244 (P) spliceosomal tri-snRNP ... GO:0000353 (P) formation of quadruple ... GO:0005688 (C) U6 snRNP GO:0046540 (C) U4/U6 x U5 tri-snRNP co... |
URS0000B21DFD | RF00026 | Saccharomyces cerevisiae 4932 |
28045380 | 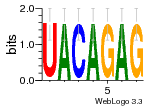 383 |
Assembly1 GOL:B:202 51,53~54 |
| 18 |
5vaj:C (1.95Å) |
11 | N/A | URS0000BC4688 | N/A | synthetic construct 32630 |
28854734 | Assembly1 GOL:C:101 8~9 |
|
| 19 |
6h9i:E (2.29Å) |
25 | N/A | URS00021ED981 | N/A | Aromatoleum aromaticum EbN1 76114 |
30397343 | Assembly2 GOL:E:101 10~11 |
|
| 20 |
6hhq:AR (3.1Å) |
3147 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000061F377 | RF02540 RF02543 |
Saccharomyces cerevisiae 4932 |
30759226 | 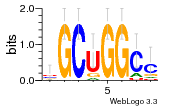 M107_0.6 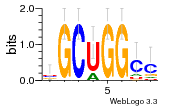 M080_0.6 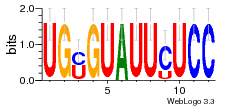 M299_0.6  16 |
Assembly2 GOL:AR:4262 1448,2351~2352 |
| 21 |
6ozl:C (2.1Å) |
15 | N/A | N/A | N/A | synthetic construct 32630 |
31444105 | Assembly1 GOL:C:101 17~18 |
|
| 22 |
6ozn:C (1.9Å) |
15 | N/A | N/A | N/A | synthetic construct 32630 |
31444105 | Assembly1 GOL:C:102 17~18 |
|
| 23 |
7k98:C (2.19Å) |
77 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000035A243 | RF00005 | Mycobacterium tuberculosis H37Rv 83332 |
33885823 | Assembly1 GOL:C:103 5~6 |
|
| 24 |
7l3r:A (1.01Å) |
27 | N/A | URS000080DEF4 | N/A | synthetic construct 32630 |
33856457 | Assembly1 GOL:A:2701 2662~2663 |
|
| 25 |
7oax:A (2.24Å) |
51 | N/A | URS0002311967 | N/A | synthetic construct 32630 |
34112799 | Assembly1 GOL:A:104 21~23,25 |
|
| 26 |
7oax:C (2.24Å) |
51 | N/A | URS0002311967 | N/A | synthetic construct 32630 |
34112799 | Assembly1 GOL:C:104 23,25 |
|
| 27 |
7qr3:C (2.18Å) |
69 | EC:3.1.-.- GO:0004540 (F) RNA nuclease activity |
URS0002617738 | RF00622 | Pan troglodytes 9598 |
N/A | Assembly1 GOL:C:101 57~58 |
|
| 28 |
7qsh:A (0.86Å) |
25 | N/A | URS000268136D | N/A | synthetic construct 32630 |
35687141 | Assembly1 GOL:A:2703 2654,2656 |
|
| 29 |
7xd8:B (2.85Å) |
12 | N/A | URS00026177B2 | N/A | synthetic construct 32630 |
36577062 | Assembly1 GOL:C:1201 1006~1007 |
|
| 30 |
7xd8:E (2.85Å) |
11 | N/A | URS00026177B2 | N/A | synthetic construct 32630 |
36577062 | Assembly2 GOL:E:1101 1005~1006 |
|
| 31 |
7xd9:E (2.58Å) |
13 | N/A | URS00026177B2 | N/A | synthetic construct 32630 |
36577062 | Assembly2 GOL:E:1101 1005~1006 GOL:E:1102 1004~1005 |
|
| 32 |
7xd9:K (2.58Å) |
16 | N/A | URS00026177B2 | N/A | synthetic construct 32630 |
36577062 | Assembly4 GOL:K:1101 1004~1005 |
|
| 33 |
8tqx:D (2.09Å) |
19 | N/A | N/A | N/A | Zika virus 64320 |
38383158 | Assembly1 GOL:D:101 13~14 |
|
| 34 |
8tsv:C (1.51Å) |
25 | N/A | N/A | N/A | Zika virus 64320 |
38383158 | Assembly2 GOL:C:101 6~7 |
|
| 35 |
8tsv:D (1.51Å) |
25 | N/A | N/A | N/A | Zika virus 64320 |
38383158 | Assembly2 GOL:D:101 3~4 |

Download all results in tab-seperated text for
35 RNAs.
Click the entry in the '#' column to view structure details of that RNA chain.
Hover over PDB to view the title of the structure.
Hover over Rfam to view names of Rfam families.
Hover over PubMed to view title of the PubMed publications.
Hover over Ligand to view the name of the ligand and the ligand-binding nucleotides on the RNA.
<<
<
1
>
>>
<<
<
1
>
>>
Reference:
