| # | PDB (Resolution) |
Length | EC number & GO term |
RNAcentral | Rfam | Taxon | PubMed | ATtRACT motif |
Ligand & binding nucleotides |
|---|---|---|---|---|---|---|---|---|---|
| 1 |
1yyk:E (2.5Å) |
12 | N/A | URS000080DE83 | N/A | synthetic construct 32630 |
16216575 | Assembly1 TRS:E:201 4~5 |
|
| 2 |
1yyo:E (2.9Å) |
12 | N/A | URS000080DE83 | N/A | synthetic construct 32630 |
16216575 | Assembly1 TRS:E:201 3~5 |
|
| 3 |
5d8h:A (2.8Å) |
74 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS0000A76356 | RF02540 | Methanocaldococcus jannaschii 2190 |
N/A | Assembly1 TRS:A:1314 1166,1210 TRS:A:1315 1168,1197,1199 |
|
| 4 |
5j5b:DA (2.8Å) |
2897 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS00004B0F34 | RF02540 RF02541 |
Escherichia coli K-12 83333 |
27382179 | Assembly1 TRS:DA:3219 2255~2256 |
|
| 5 |
5j91:DA (2.96Å) |
2897 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS00004B0F34 | RF02540 RF02541 |
Escherichia coli K-12 83333 |
27382179 | Assembly1 TRS:DA:3219 2255~2256 |
|
| 6 |
6du4:B (1.7Å) |
29 | N/A | URS0000D7791F | N/A | Homo sapiens 9606 |
30197297 | Assembly1 TRS:B:101 22~23 |
|
| 7 |
7r97:C (1.804Å) |
28 | N/A | URS000080DE20 | N/A | Escherichia coli 562 |
35829618 30916338 |
Assembly1 TRS:C:103 11~12 |
|
| 8 |
7r97:D (1.804Å) |
28 | N/A | URS000080DE20 | N/A | Escherichia coli 562 |
35829618 30916338 |
Assembly1 TRS:D:103 11~12 |
|
| 9 |
8glp:L5 (1.67Å) |
3546 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS00026813F0 | RF02543 | Homo sapiens 9606 |
37020024 |  M054_0.6  1306  769  706 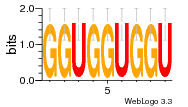 1303 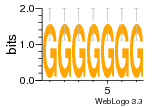 762 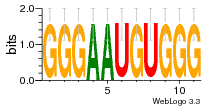 872 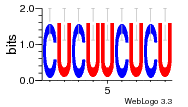 1148 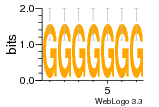 1149 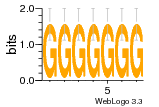 764 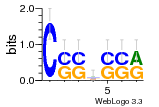 167 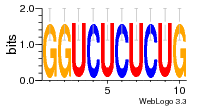 21 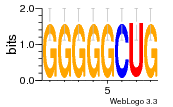 930 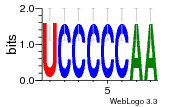 1029 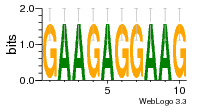 1075 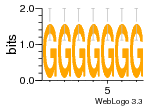 1072 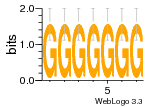 1073 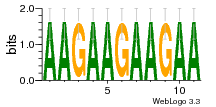 s77 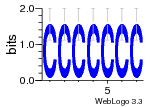 1071 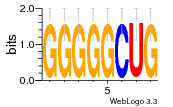 819  1373  938  1375  s52  s50  931  M272_0.6  936 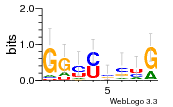 s59 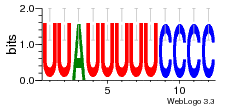 995 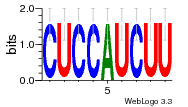 994 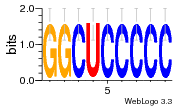 1314 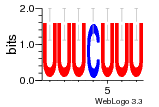 991 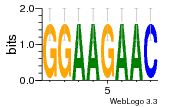 1312 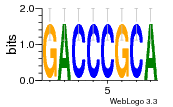 1311 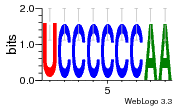 1028  1150 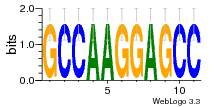 123 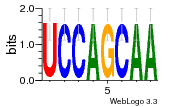 124 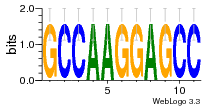 125 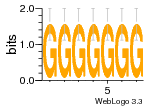 1068  s72  1060  s100  s102 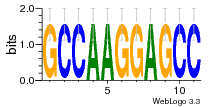 52 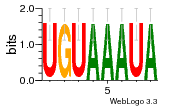 533 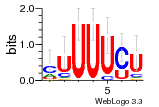 M227_0.6 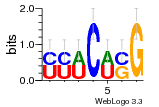 1384 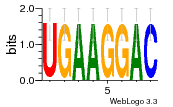 375 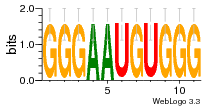 926 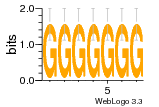 822 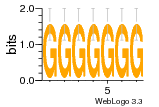 1128 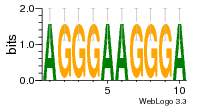 988 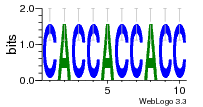 1321 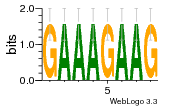 1083 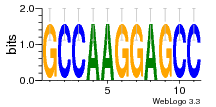 705 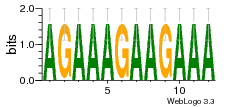 1328 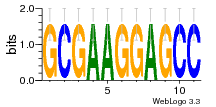 707 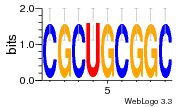 1124 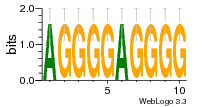 987 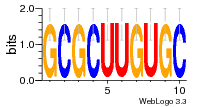 1126 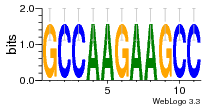 703 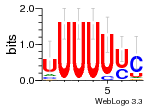 M077_0.6 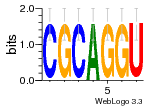 1237 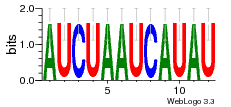 83 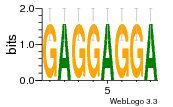 1181 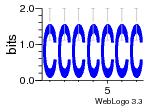 797 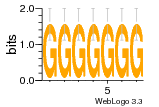 796 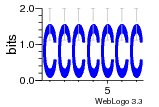 795 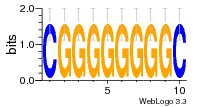 836 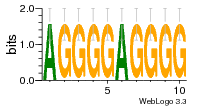 837 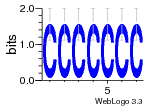 791  790  838  1331  s70  1016 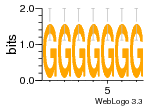 1017 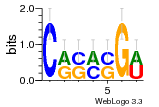 1393 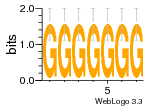 610 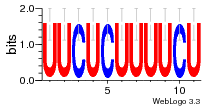 1012 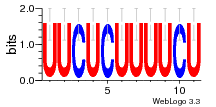 1013 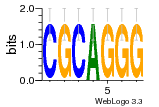 1236 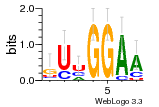 M290_0.6 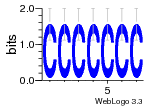 763 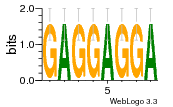 1231 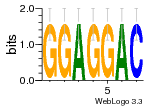 1232 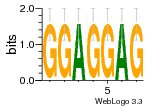 1233 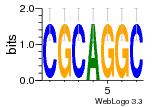 1335  1334  M109_0.6  1336  1238  1239  1333  589  776 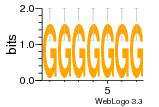 s96 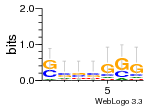 s99 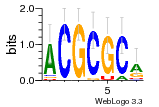 M273_0.6 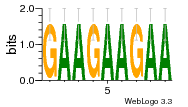 1389 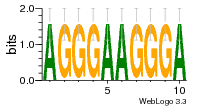 902 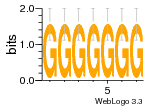 821 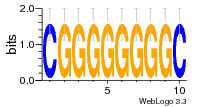 900 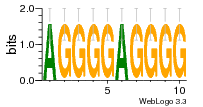 901 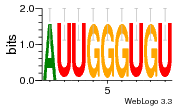 906 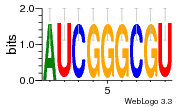 905 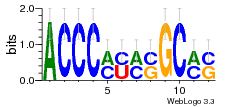 33 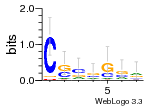 s48  s17  970  M044_0.6  1245  s62  s61 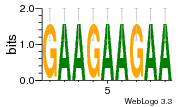 1227 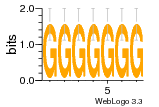 883 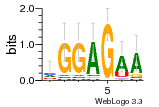 M035_0.6 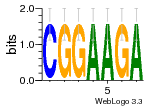 1223 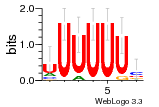 M156_0.6 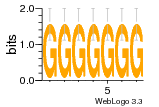 604 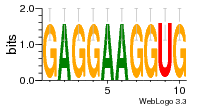 1347 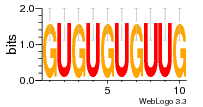 60  s81  M349_0.6  595  977 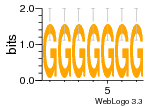 976 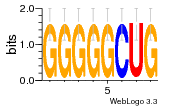 974 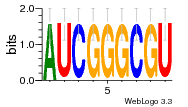 852 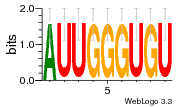 853  M154_0.6  884  s55  979  M153_0.6  183  185  546 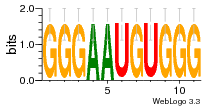 815 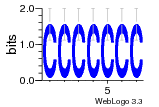 1036 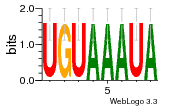 503 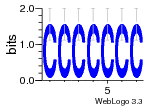 1034 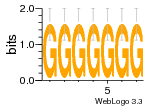 1035 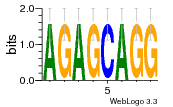 1212 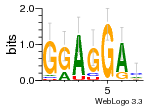 M102_0.6 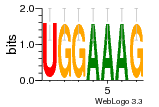 1216 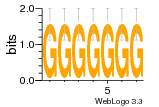 1042 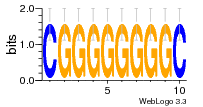 986 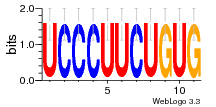 467 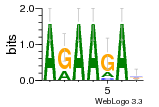 M148_0.6 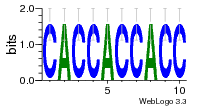 1354 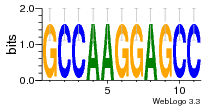 708  M103_0.6 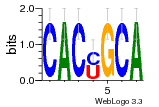 169 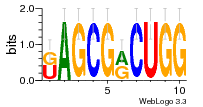 91  1127  M070_0.6  1026  M020_0.6 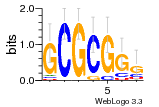 M050_0.6 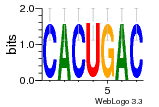 1187 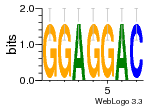 1182 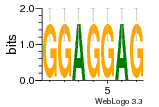 1183  1037  727  M347_0.6  722  935 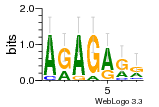 M086_0.6 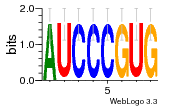 1164 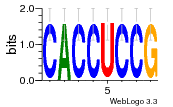 1165 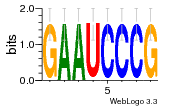 1166  606  886  156  748  749  608  M061_0.6  958  1055 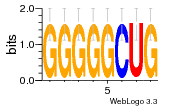 878 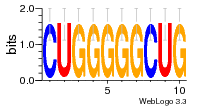 879 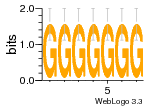 1059 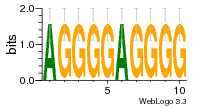 953 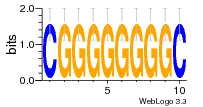 952 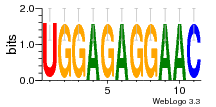 42 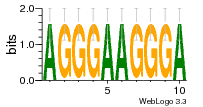 954 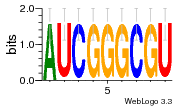 957 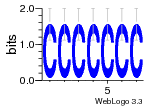 1107 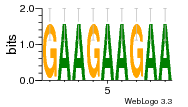 1352 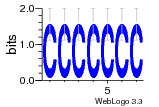 774 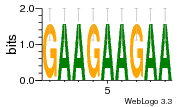 1215 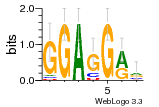 M104_0.6 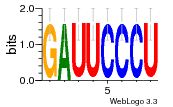 1173 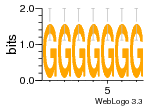 770 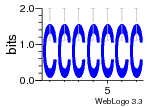 773 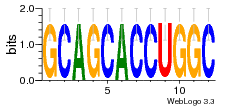 1271 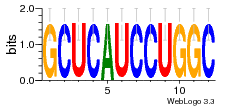 1272 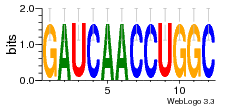 1273 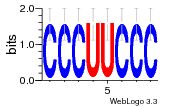 779 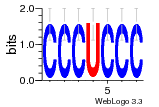 778  1178 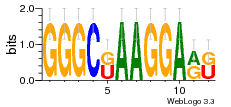 78 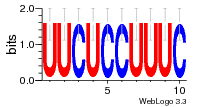 1285  M083_0.6 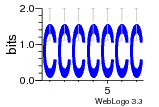 1041 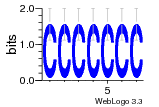 1040 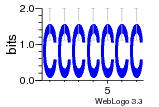 1043 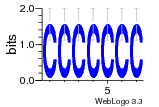 1045 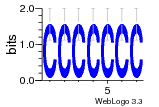 1044  1288  354  1360  684  800  1368 |
Assembly1 TRS:L5:5124 4587~4588,4711~4712 TRS:L5:5125 714~715,1280 |

Download all results in tab-seperated text for
9 RNAs.
Click the entry in the '#' column to view structure details of that RNA chain.
Hover over PDB to view the title of the structure.
Hover over Rfam to view names of Rfam families.
Hover over PubMed to view title of the PubMed publications.
Hover over Ligand to view the name of the ligand and the ligand-binding nucleotides on the RNA.
<<
<
1
>
>>
<<
<
1
>
>>
Reference:
