| # | PDB (Resolution) |
Length | EC number & GO term |
RNAcentral | Rfam | Taxon | PubMed | ATtRACT motif |
Ligand & binding nucleotides |
|---|---|---|---|---|---|---|---|---|---|
| 1 |
1a34:B (1.81Å) |
10 | N/A | URS000080DEB3 | N/A | synthetic construct 32630 |
9514737 | Assembly1 U:B:11 8~9 |
|
| 2 |
1a34:C (1.81Å) |
10 | N/A | URS00002C5188 | N/A | synthetic construct 32630 |
9514737 | Assembly1 U:B-6:11 8~9 |
|
| 3 |
7pd3:A (3.4Å) |
1280 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS0002311A1E | RF02541 RF02540 |
Homo sapiens 9606 |
34609277 | 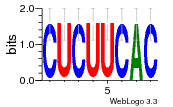 591 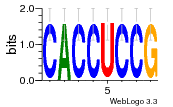 1165 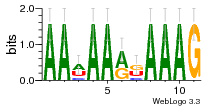 M269_0.6 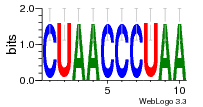 156  M013_0.6 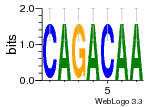 1228 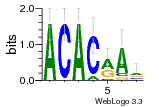 M089_0.6 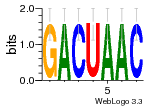 1199 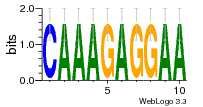 693 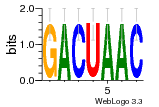 1191 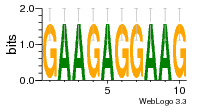 1075  M111_0.6  1271 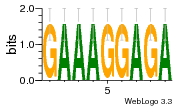 1395 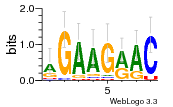 M272_0.6 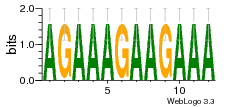 1328 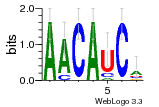 M081_0.6 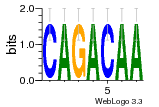 1310 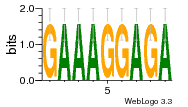 1353 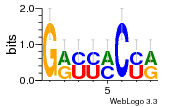 1334 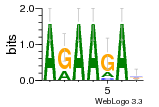 M148_0.6 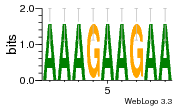 1239 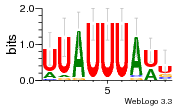 M350_0.6 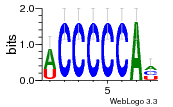 s14 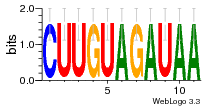 74 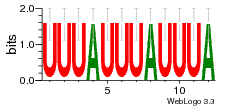 161 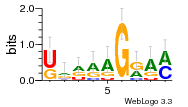 s25  1289 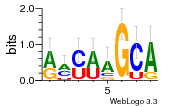 s44 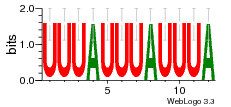 1100 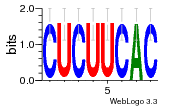 1246 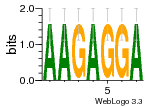 1385 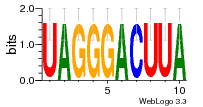 687 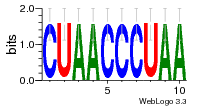 800 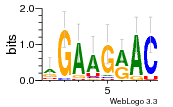 s60 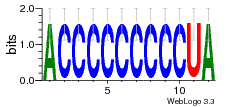 s61 |
Assembly1 U:A:3315 3000~3001 U:A:3322 2714,2997 U:A:3345 1962,2502 U:A:3347 1956,2720~2721 U:A:3349 2312,2676~2677 U:A:3353 2685~2686,2715~2716 U:A:3355 2711~2712 |
| 4 |
8evr:B5 (2.87Å) |
1173 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS0002681484 | RF01960 RF02542 RF01959 RF00177 |
Saccharomyces cerevisiae 4932 |
37595043 | 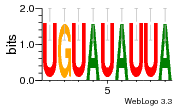 101 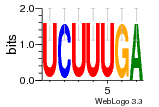 441 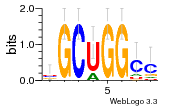 M080_0.6 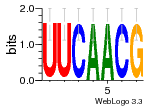 386 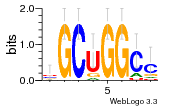 M107_0.6 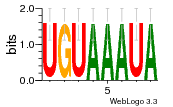 497  490 |
Assembly1 U:BC:306 13,1086~1087 U:BO:201 992,1006 U:B5:1850 1167~1168 U:B5:1851 1166~1167 U:B5:1854 1158,1163 U:B5:1860 1157,1160 U:B5:1865 1152~1153 U:B5:1866 1151~1152 U:B5:1867 1149,1151 U:B5:1917 1005~1006 |
| 5 |
8evs:A1 (2.62Å) |
3198 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS0002681472 | RF02543 | Saccharomyces cerevisiae 4932 |
37595043 |  M107_0.6  M080_0.6  M299_0.6  16 |
Assembly1 U:A1:3407 2124~2125 U:A1:3423 2255,2262 |
| 6 |
8evs:B5 (2.62Å) |
1173 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS0002681484 | RF01960 RF02542 RF01959 RF00177 |
Saccharomyces cerevisiae 4932 |
37595043 |  101  441  M080_0.6  386  M107_0.6  497  490 |
Assembly1 U:BO:201 992,1006 U:BT:214 1171~1172 U:B5:1821 13,1086~1087 U:B5:1831 1188~1189 U:B5:1852 1167~1168 U:B5:1853 1166~1167 U:B5:1856 1158,1163 U:B5:1858 1164~1165 U:B5:1867 1152~1153 U:B5:1868 1151~1152 U:B5:1869 1149,1151 U:B5:1920 1005~1006 U:An:111 978~979 U:EC:7006 1001~1002 |
| 7 |
8evs:EC (2.62Å) |
195 | N/A | URS000268147B | N/A | Saccharomyces cerevisiae 4932 |
37595043 | Assembly1 U:EC:7006 6950~6951 |
|
| 8 |
8evt:A1 (2.2Å) |
3192 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS0002681478 | RF02543 | Saccharomyces cerevisiae 4932 |
37595043 |  M107_0.6  M080_0.6  M299_0.6  16 |
Assembly1 U:A1:3405 2257~2258 U:A1:3408 2124~2125 |
| 9 |
8evt:B5 (2.2Å) |
1173 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS0002681484 | RF01960 RF02542 RF01959 RF00177 |
Saccharomyces cerevisiae 4932 |
37595043 |  101  441  M080_0.6  386  M107_0.6  497  490 |
Assembly1 U:BC:305 1144~1145 U:BT:212 1171~1172 U:B5:1823 13,1086~1087 U:B5:1856 1166~1167 U:B5:1859 1158,1163 U:B5:1865 1157,1160 U:B5:1870 1152~1153 U:B5:1871 1151~1152 U:B5:1872 1149,1151 U:B5:1925 1005~1006 U:B5:1933 992,1006 |

Download all results in tab-seperated text for
9 RNAs.
Click the entry in the '#' column to view structure details of that RNA chain.
Hover over PDB to view the title of the structure.
Hover over Rfam to view names of Rfam families.
Hover over PubMed to view title of the PubMed publications.
Hover over Ligand to view the name of the ligand and the ligand-binding nucleotides on the RNA.
<<
<
1
>
>>
<<
<
1
>
>>
Reference:
