| # | PDB (Resolution) |
Length | EC number & GO term |
RNAcentral | Rfam | Taxon | PubMed | ATtRACT motif |
Ligand & binding nucleotides |
|---|---|---|---|---|---|---|---|---|---|
| 1 |
7zs5:1 (3.2Å) |
3248 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000061F377 | RF02543 | Saccharomyces cerevisiae 4932 |
36801861 | 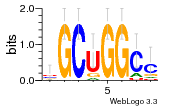 M107_0.6 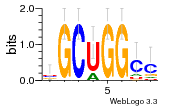 M080_0.6 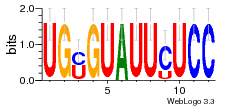 M299_0.6 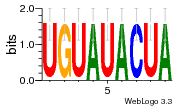 16 |
Assembly1 UNK:1:3412 2403~2404 |
| 2 |
8wkp:A16S (4.62Å) |
1325 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
N/A | RF01959 RF00177 RF01960 RF02542 |
Sulfolobus acidocaldarius DSM 639 330779 |
38599770 | Assembly1 UNK:A16S:1602 829~830 UNK:A16S:1603 794,829~830 |

Download all results in tab-seperated text for
2 RNAs.
Click the entry in the '#' column to view structure details of that RNA chain.
Hover over PDB to view the title of the structure.
Hover over Rfam to view names of Rfam families.
Hover over PubMed to view title of the PubMed publications.
Hover over Ligand to view the name of the ligand and the ligand-binding nucleotides on the RNA.
<<
<
1
>
>>
<<
<
1
>
>>
Reference:
