| # | PDB (Resolution) |
Length | EC number & GO term |
RNAcentral | Rfam | Taxon | PubMed | ATtRACT motif |
Ligand & binding nucleotides |
|---|---|---|---|---|---|---|---|---|---|
| 1101 |
4v8z:B2 (6.6Å) |
1781 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000080DE56 | RF01960 RF01959 RF00177 |
Saccharomyces cerevisiae 4932 |
24200810 |  101  441  M080_0.6  386 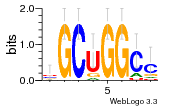 M107_0.6 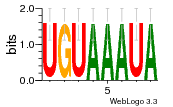 497 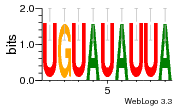 490 |
Assembly1 OHX:AI:301 118,293 OHX:B2:1901 1642,1756~1758 OHX:B2:1902 984~986,1014 OHX:B2:1903 1414,1418 OHX:B2:1904 549~550,583~584 OHX:B2:1905 104,353,355 OHX:B2:1906 1085,1298 OHX:B2:1907 1212~1213,1449 OHX:B2:1908 1046~1047 OHX:B2:1909 1497~1498,1508 OHX:B2:1910 867~868 OHX:B2:1912 129,264 OHX:B2:1913 992,1006 OHX:B2:1914 103,355~356 OHX:B2:1915 833,837 OHX:B2:1916 120,452~453 OHX:B2:1917 565,577 OHX:B2:1918 42,438 OHX:B2:1919 1661~1662,1738 OHX:B2:1920 405,423 OHX:B2:1922 1672,1727 OHX:B2:1923 980,1015 OHX:B2:1924 57~58 OHX:B2:1925 1237~1239 OHX:B2:1926 120,122 OHX:B2:1927 976,1023 OHX:B2:1930 523,527 OHX:B2:1931 800,802 OHX:B2:1932 1667~1668 OHX:B2:1933 32~33 OHX:B2:1936 1486,1520,1522 OHX:B2:1937 357~358 OHX:B2:1938 760~761 OHX:B2:1941 301,384 OHX:B2:1942 1155,1314 OHX:B2:1943 8,1139~1140 OHX:B2:1944 62,82~83 OHX:B2:1945 205,261 OHX:B2:1947 1110~1111 OHX:B2:1948 515,530 OHX:B2:1949 1413,1417 OHX:B2:1950 51~52 OHX:B2:1951 131,176 OHX:B2:1952 1114,1129~1130 OHX:B2:1953 1495,1596,1598 OHX:B2:1954 470~471 OHX:B2:1955 1294~1295 OHX:B2:1957 108~110 OHX:B2:1958 877~878 OHX:B2:1959 1718,1720 OHX:B2:1961 25,27,40 OHX:B2:1962 1317~1318 OHX:B2:1963 1283~1284 OHX:B2:1964 1653~1654,1745 OHX:B2:1965 938,940 OHX:B2:1966 101,358 OHX:B2:1968 990~991,1011 OHX:B2:1969 321~322 OHX:B2:1970 1764~1766,1770,1794 OHX:B2:1971 245,248 OHX:B2:1972 894,915 OHX:B2:1973 1353,1368~1369 OHX:B2:1974 698,738~740 OHX:B2:1976 1725~1726 OHX:B2:1977 1485~1486 OHX:B2:1978 205~206 OHX:B2:1982 1747~1748 OHX:B2:1983 364~365 OHX:B2:1984 329,336 OHX:B2:1985 422~423 OHX:B2:1987 1366~1367 OHX:B2:1988 1679~1680 OHX:B2:1990 1201~1202,1599 OHX:B2:1992 1150,1637 OHX:B2:1994 1159,1193,1282~1283,1285 OHX:B2:1995 213~214 OHX:B2:1996 1167~1168,1588 OHX:B2:1997 1181~1182 OHX:B2:1998 21~22,598 OHX:B2:1999 1370,1518 OHX:B2:2000 880,1018 OHX:B2:2002 1669~1670 OHX:B2:2005 318,346 OHX:B2:2007 348~349 OHX:B2:2009 701~702,732 OHX:B2:2010 383~384 OHX:B2:2011 991,1010~1011 OHX:B2:2012 1157~1158,1616 OHX:B2:2016 982~983 OHX:B2:2018 1665,1734~1735 OHX:B2:2019 764,769~770 OHX:B2:2020 1589~1590 OHX:B2:2022 833,835 OHX:B2:2023 562,579~580 OHX:B2:2024 1351~1353 OHX:B2:2025 1282~1283 OHX:B2:2026 461~462 OHX:B2:2028 1005~1006,1769 OHX:B2:2029 1500,1507 OHX:B2:2031 1117~1119 OHX:B2:2033 1152~1153 OHX:B2:2035 1175,1561 OHX:B2:2036 1041~1042 OHX:B2:2037 73,76~77,80 OHX:B2:2038 687~688 OHX:B2:2039 313~314 OHX:B2:2040 1233,1245~1247 OHX:B2:2042 1035,1095 OHX:B2:2044 1537,1570~1572 OHX:B2:2047 434,437 OHX:B2:2048 1303,1305 OHX:B2:2051 622~624 OHX:B2:2057 367,759 OHX:B2:2059 1051,1053 OHX:B2:2060 1142,1631 OHX:B2:2061 1469,1589 OHX:B2:2062 430~431,438 OHX:B2:2064 1336~1337,1381,1485 OHX:B2:2067 1590~1591 OHX:B2:2068 1634,1639 OHX:B2:2069 222~223,837~840 OHX:B2:2071 1618~1620 OHX:B2:2072 885,925 OHX:B2:2075 1055~1056,1062~1064 OHX:B2:2078 1310~1311,1314 OHX:B2:2079 792~793 OHX:B2:2081 1214~1215,1446,1448 OHX:B5:3402 1004,1780 |
| 1102 |
4v8z:B5 (6.6Å) |
3147 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000061F377 | RF02540 RF02543 |
Saccharomyces cerevisiae 4932 |
24200810 |  M107_0.6  M080_0.6 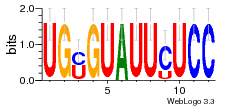 M299_0.6  16 |
Assembly1 OHX:B5:3402 2263~2264 |
| 1103 |
4v8z:B7 (6.6Å) |
121 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000055688D | RF00001 | Saccharomyces cerevisiae 4932 |
24200810 | Assembly1 OHX:B7:201 75,77 OHX:B7:202 14,58~59 OHX:B7:206 109~110 OHX:B7:219 79,81 |
|
| 1104 |
4v97:AA (3.516Å) |
1504 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000080E306 | RF00177 RF01959 RF01960 |
Thermus thermophilus HB8 300852 |
23630274 | Assembly1 PAR:AA:7111 1405,1408,1490~1495 |
|
| 1105 |
4v97:CA (3.516Å) |
1504 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000080E306 | RF00177 RF01959 RF01960 |
Thermus thermophilus HB8 300852 |
23630274 | Assembly2 PAR:CA:1741 1405,1407~1408,1490~1494 |
|
| 1106 |
4v9a:AA (3.2999Å) |
1506 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000080DEDF | RF00177 RF01959 RF01960 |
Thermus thermophilus HB8 300852 |
23431179 | Assembly1 TAC:AA:1833 1054,1195~1198 |
|
| 1107 |
4v9a:CA (3.2999Å) |
1506 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000080DEDF | RF00177 RF01959 RF01960 |
Thermus thermophilus HB8 300852 |
23431179 | Assembly2 TAC:CA:1805 1054,1195,1197~1198 |
|
| 1108 |
4v9b:AA (3.1Å) |
1506 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000080DEDF | RF00177 RF01959 RF01960 |
Thermus thermophilus HB8 300852 |
23431179 | Assembly1 T1C:AA:1837 1054,1195~1198 |
|
| 1109 |
4v9b:CA (3.1Å) |
1506 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000080DEDF | RF00177 RF01959 RF01960 |
Thermus thermophilus HB8 300852 |
23431179 | Assembly2 T1C:CA:1800 966,1054,1195,1197~1198 |
|
| 1110 |
4v9c:AA (3.3Å) |
1539 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS00000ABFE9 | RF00177 RF01959 RF01960 |
Escherichia coli K-12 83333 |
22902368 | Assembly1 NMY:AA:1655 1405,1407~1408,1490~1495 NMY:AA:1656 1417,1419~1420,1477 NMY:AA:1657 902,1513~1515 |
|
| 1111 |
4v9c:BA (3.3Å) |
2897 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS00004B0F34 | RF02540 RF02541 |
Escherichia coli K-12 83333 |
22902368 | Assembly1 NMY:BA:3161 1906,1921~1923 NMY:BA:3162 990~992,1157~1158,1162~1164,11 84~1185 NMY:BA:3163 1202,1204~1206,1239~1240 NMY:BA:3164 482,492,494,507~508 NMY:BA:3165 2674,2727~2728,2730~2731 NMY:BA:3166 536~540,550~551,553 NMY:BA:3167 1858~1859,1861,1880,1882~1883 |
|
| 1112 |
4v9c:CA (3.3Å) |
1538 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS00000ABFE9 | RF00177 RF01959 RF01960 |
Escherichia coli K-12 83333 |
22902368 | Assembly2 NMY:CA:1672 1406~1408,1489~1491,1493~1494 |
|
| 1113 |
4v9c:DA (3.3Å) |
2897 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS00004B0F34 | RF02540 RF02541 |
Escherichia coli K-12 83333 |
22902368 | Assembly2 NMY:DA:3184 1921~1923,1929~1931 NMY:DA:3185 2673~2674,2727~2731 NMY:DA:3186 536~540 NMY:DA:3187 1858~1859,1879~1880,1882 NMY:DA:3188 991~992,1157~1159,1162~1163,11 84 NMY:DA:3189 482,488,492~494,507 NMY:DA:3190 1204~1205,1238~1240 |
|
| 1114 |
4v9f:0 (2.404Å) |
2808 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000080E02E | RF02540 RF00002 |
Haloarcula marismortui ATCC 43049 272569 |
23695244 10937989 |
Assembly1 CL:0:8185 1300,1329 CL:0:8190 1072,1087,1291 ACY:0:8212 2756,2895~2897 ACY:0:8213 966~967,1002 |
|
| 1115 |
4v9q:AA (3.4Å) |
2832 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000080DD88 | RF02540 RF02541 RF00002 |
Thermus thermophilus HB27 262724 |
23824292 | Assembly1 BLS:AA:4001 2063~2064,2251~2252,2439,2600~ 2601 |
|
| 1116 |
4v9q:BV (3.4Å) |
77 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000041AF00 | RF00005 | Alcaligenes sp. 512 |
23824292 | Assembly1 BLS:AA:4001 74~76 |
|
| 1117 |
4v9q:CA (3.4Å) |
2832 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000080DD88 | RF02540 RF02541 RF00002 |
Thermus thermophilus HB27 262724 |
23824292 | Assembly2 BLS:CA:4405 2063~2064,2251~2252,2439,2600~ 2601 |
|
| 1118 |
4v9q:DV (3.4Å) |
77 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000041AF00 | RF00005 | Alcaligenes sp. 512 |
23824292 | Assembly2 BLS:CA:4405 74~76 |
|
| 1119 |
4v9r:BA (3.0Å) |
2731 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS00001698E8 | RF02540 RF02541 RF00002 |
Thermus thermophilus 274 |
24412368 | Assembly1 FME:AX:101 2085,2463,2597 |
|
| 1120 |
4v9r:DA (3.0Å) |
2714 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS00001698E8 | RF02540 RF02541 RF00002 |
Thermus thermophilus 274 |
24412368 | Assembly2 FME:CX:101 2451,2585 |
|
| 1121 |
4v9s:BA (3.1Å) |
2731 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS00001698E8 | RF02540 RF02541 RF00002 |
Thermus thermophilus HB8 300852 |
24412368 | Assembly1 FME:AX:101 2085,2463,2597 |
|
| 1122 |
4v9s:DA (3.1Å) |
2714 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS00001698E8 | RF02540 RF02541 RF00002 |
Thermus thermophilus HB8 300852 |
24412368 | Assembly2 FME:CX:101 2063,2451,2585 |
|
| 1123 |
4w29:AA (3.8Å) |
1511 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000080DFCE | RF00177 RF01959 RF01960 |
Thermus thermophilus HB27 262724 |
25190797 | Assembly1 NMY:AA:1601 1405~1406,1408,1491~1494 |
|
| 1124 |
4w29:BA (3.8Å) |
2879 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000080DEC7 | RF02540 RF02541 RF00002 |
Thermus thermophilus HB27 262724 |
25190797 | Assembly1 NMY:BA:2902 1831~1832,1933,1974 NMY:BA:2903 1963~1966 NMY:BA:2904 1905,1930,1966~1972,2593 |
|
| 1125 |
4w29:CA (3.8Å) |
1511 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000080DFCE | RF00177 RF01959 RF01960 |
Thermus thermophilus HB27 262724 |
25190797 | Assembly2 NMY:CA:1601 1405~1406,1408,1491~1494 |
|
| 1126 |
4w29:DA (3.8Å) |
2879 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000080DEC7 | RF02540 RF02541 RF00002 |
Thermus thermophilus HB27 262724 |
25190797 | Assembly2 NMY:DA:2901 1831~1832,1933~1934,1974 |
|
| 1127 |
4w2e:A (2.9Å) |
2873 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS00001698E8 | RF02540 RF02541 RF00002 |
Thermus thermophilus 274 |
25104389 | Assembly1 PHE:w:107 2451~2452,2585 |
|
| 1128 |
4w2f:AA (2.4Å) |
1497 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000080DFED | RF00177 RF01959 RF01960 |
Thermus thermophilus HB8 300852 |
25306919 | Assembly1 UAM:AA:3232 693,788,794~796,1506 |
|
| 1129 |
4w2f:AV (2.4Å) |
13 | N/A | N/A | N/A | synthetic construct 32630 |
25306919 | Assembly1 UAM:AA:3232 14~15 |
|
| 1130 |
4w2f:CA (2.4Å) |
1503 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000080DFED | RF00177 RF01959 RF01960 |
Thermus thermophilus HB8 300852 |
25306919 | Assembly2 UAM:CA:3202 693,788,792,794~796,1506 |
|
| 1131 |
4w2g:AA (2.55Å) |
1498 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000080DFED | RF00177 RF01959 RF01960 |
Thermus thermophilus HB8 300852 |
25306919 | Assembly1 PCY:AA:3231 693,787~788,795~796 |
|
| 1132 |
4w2g:AY (2.55Å) |
74 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS00005AA258 | RF00005 | Escherichia coli 562 |
25306919 | Assembly1 PCY:AA:3231 34~35 |
|
| 1133 |
4w2g:CA (2.55Å) |
1503 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000080DFED | RF00177 RF01959 RF01960 |
Thermus thermophilus HB8 300852 |
25306919 | Assembly2 PCY:CA:3178 693,787~788,795~796 |
|
| 1134 |
4w2h:AA (2.7Å) |
1497 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000080DFED | RF00177 RF01959 RF01960 |
Thermus thermophilus HB8 300852 |
25306919 | Assembly1 PCY:AA:3191 693,788~789,795~796 |
|
| 1135 |
4w2h:CA (2.7Å) |
1503 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000080DFED | RF00177 RF01959 RF01960 |
Thermus thermophilus HB8 300852 |
25306919 | Assembly2 PCY:CA:3176 693~694,787~788,795~796 |
|
| 1136 |
4w2i:AA (2.7Å) |
1496 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000080DFED | RF00177 RF01959 RF01960 |
Thermus thermophilus HB8 300852 |
25306922 | Assembly1 NEG:AA:3216 965~966,1054,1197~1198 NEG:AA:3217 569,873 NEG:AA:3218 941~942,1336 NEG:AA:3219 669~672,807 |
|
| 1137 |
4w2i:BA (2.7Å) |
2822 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS00001698E8 | RF02540 RF02541 RF00002 |
Thermus thermophilus HB8 300852 |
25306922 | Assembly1 NEG:AA:3220 1966~1969,1976~1977 NEG:AA:3221 785~786,801~802,804 NEG:AA:3222 553~554,558~559,580~581 NEG:AW:3004 1963~1966,2616 NEG:AX:3013 2333~2336,2342~2344 |
|
| 1138 |
4w2i:CA (2.7Å) |
1503 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000080DFED | RF00177 RF01959 RF01960 |
Thermus thermophilus HB8 300852 |
25306922 | Assembly2 NEG:CA:3170 965,1054,1197~1198 NEG:CA:3171 569~570,873 NEG:CA:3172 942,1336 NEG:CA:3173 669~672 |
|
| 1139 |
4w2i:DA (2.7Å) |
2800 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS00001698E8 | RF02540 RF02541 RF00002 |
Thermus thermophilus HB8 300852 |
25306922 | Assembly2 NEG:CA:3174 1945~1946,1949~1950,1954~1955 NEG:CA:3175 739,754~755,757 NEG:CA:3176 528~529,533,557~559 NEG:CA:3177 13~16,524~525 NEG:CW:3002 1941~1942,1944,2604 NEG:CX:3004 2321~2324,2329~2332 |
|
| 1140 |
4w90:C (3.118Å) |
101 | GO:0180001 (F) cyclic-di-AMP binding GO:0010468 (P) regulation of gene expr... |
URS000080DDC9 | RF00379 | Bacillus subtilis 1423 |
25271255 | Assembly1 2BA:C:201 9,25~28,60~62,95~96,100,109~11 0 2BA:C:202 5~7,64,80~82,89~90,114~116 |
|
| 1141 |
4w92:C (3.209Å) |
96 | GO:0180001 (F) cyclic-di-AMP binding GO:0010468 (P) regulation of gene expr... |
URS000080DDC9 | RF00379 | Bacillus subtilis 1423 |
25271255 | Assembly1 EDO:C:207 23~24,110 2BA:C:208 5~7,64,80~82,89~90,114~116 2BA:C:209 9,25~28,60~63,95~96,100,109~11 0 |
|
| 1142 |
4wce:X (3.526Å) |
2708 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS00001450D9 | RF02540 RF02541 |
Staphylococcus aureus subsp. aureus NCTC 8325 93061 |
26464510 | Assembly1 MPD:X:3001 1604~1605 MPD:X:3002 2549~2550 MPD:X:3003 2088~2089,2470~2471 MPD:X:3004 204,207 MPD:X:3005 1340~1341,1650~1651 MPD:X:3006 691~692 MPD:X:3007 627~628,1289 MPD:X:3008 197~198,712~713 MPD:X:3011 622,1298 EPE:X:3311 2065~2066,2068 SPD:X:3313 1019~1020 SPD:X:3315 2571,2669~2672 |
|
| 1143 |
4wf1:CA (3.09Å) |
1539 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000080DDC0 | RF00177 RF01959 RF01960 |
Escherichia coli str. K-12 substr. MG1655 511145 |
25368144 | Assembly2 NEG:CA:1657 965,1054,1196~1198 |
|
| 1144 |
4wf9:X (3.427Å) |
2712 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS00001450D9 | RF02540 RF02541 |
Staphylococcus aureus subsp. aureus NCTC 8325 93061 |
26464510 | Assembly1 TEL:X:3001 791,2085~2086,2532,2638 MPD:X:3002 1560~1561,1604~1605 MPD:X:3003 2708,2711~2713,2747 MPD:X:3004 1316~1317 MPD:X:3005 2059,2082,2480,2526 MPD:X:3006 508~509 MPD:X:3007 843~845 MPD:X:3008 513~514 MPD:X:3009 572,2807 SPD:X:3362 2478~2479 SPD:X:3363 2571,2669~2670,2672 SPD:X:3364 1975~1978 SPD:X:3365 373,522~523,1248,1275 EOH:X:3368 1384~1385 |
|
| 1145 |
4wfa:X (3.392Å) |
2711 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS00001450D9 | RF02540 RF02541 |
Staphylococcus aureus subsp. aureus NCTC 8325 93061 |
26464510 | Assembly1 ZLD:X:3001 2088,2474,2478~2479,2528,2531~ 2533,2612 MPD:X:3003 1316~1317 MPD:X:3005 1560~1561,1605 MPD:X:3006 2740,2744~2746 MPD:X:3007 350,1249~1250 MPD:X:3009 635~637,707 EPE:X:3423 789~791,2637~2639 EPE:X:3424 1698~1700 EPE:X:3425 2289~2290,2292~2293,2300 EPE:X:3426 626~628,1289 SPD:X:3427 1015,1018~1020,1022,1032~1033 SPD:X:3428 2669~2670,2672 SPD:X:3430 513~514 SPD:X:3432 622,1297~1299 SPD:X:3433 638~640,705 SPD:X:3434 1247,1249 EOH:X:3437 691,693 |
|
| 1146 |
4wfb:X (3.43Å) |
2707 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS00001450D9 | RF02540 RF02541 |
Staphylococcus aureus subsp. aureus NCTC 8325 93061 |
26464510 | Assembly1 3LK:X:3001 2088,2090,2478~2479,2531~2533, 2612 MPD:X:3003 15~16 MPD:X:3004 884,981 MPD:X:3005 884,982~984 MPD:X:3006 642,702 MPD:X:3007 1315~1317 MPD:X:3009 695~696 MPD:X:3012 1734,2005 MPD:X:3016 626~628,1289 EPE:X:3421 790~791,2638~2639 EPE:X:3422 1822~1823,1849 EPE:X:3423 2651,2654,2804 EPE:X:3424 1073~1075 SPD:X:3425 508,1655 SPD:X:3426 3,2657~2659,2913 SPD:X:3427 2669~2672 SPD:X:3428 1021~1022,1025 SPD:X:3429 638~639 SPD:X:3430 637,707 SPD:X:3431 2431~2432 SPD:X:3432 1340,1650~1652 SPD:X:3434 198~200,849~850 |
|
| 1147 |
4wfl:A (2.49Å) |
107 | GO:0006617 (P) SRP-dependent cotransla... GO:0048501 (C) signal recognition part... |
URS000080DF51 | RF01854 | Bacillus subtilis subsp. subtilis str. 168 224308 |
25270875 | Assembly1 NCO:A:202 67~68 NCO:A:205 18,28 NCO:A:206 57~58 NCO:A:208 81~82 NCO:A:209 106~108 NCO:A:212 3,103 NCO:A:213 73~74 NCO:A:214 35,94 NCO:A:217 21,23 NCO:A:218 11,62 |
|
| 1148 |
4wfm:A (3.1Å) |
103 | GO:0006617 (P) SRP-dependent cotransla... GO:0048501 (C) signal recognition part... |
URS000080E098 | RF01854 | Bacillus subtilis subsp. subtilis str. 168 224308 |
25270875 | Assembly1 NCO:A:202 57~58 NCO:A:203 18,28~29 NCO:A:204 11,62 NCO:A:206 73~74,88 NCO:A:207 79~80 NCO:A:209 3,103 NCO:A:212 53~54 |
|
| 1149 |
4wfm:B (3.1Å) |
103 | GO:0006617 (P) SRP-dependent cotransla... GO:0048501 (C) signal recognition part... |
URS000080E098 | RF01854 | Bacillus subtilis subsp. subtilis str. 168 224308 |
25270875 | Assembly2 NCO:B:201 57~58 NCO:B:202 11,62 NCO:B:203 7,64 NCO:B:205 17~18,28~29 NCO:B:206 73~74,88 NCO:B:207 3,103~104 NCO:B:210 16,68 |
|
| 1150 |
4wfn:X (3.54Å) |
2680 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000080DEF1 | RF02540 RF02541 |
Deinococcus radiodurans R1 = ATCC 13939 = DSM 20539 243230 |
28689968 | Assembly1 ERY:X:2902 759,2041,2484,2589~2590 |
|
| 1151 |
4woi:AA (3.0Å) |
1538 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS00000ABFE9 | RF00177 RF01959 RF01960 |
Escherichia coli str. K-12 substr. MDS42 1110693 |
26224058 | Assembly1 PAR:AA:1672 1405,1407~1408,1490~1491,1493~ 1494 |
|
| 1152 |
4woi:BA (3.0Å) |
2897 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS00004B0F34 | RF02540 RF02541 |
Escherichia coli str. K-12 substr. MDS42 1110693 |
26224058 | Assembly1 PAR:BA:3001 1906,1921~1922,1929~1930 PAR:BA:3002 2673~2674,2727~2731 PAR:BA:3003 536~538,550~551 PAR:BA:3004 482~483,492,494~495,507 PAR:BA:3005 1858~1862,1880~1882 |
|
| 1153 |
4woi:CA (3.0Å) |
2897 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS00004B0F34 | RF02540 RF02541 |
Escherichia coli str. K-12 substr. MDS42 1110693 |
26224058 | Assembly2 PAR:CA:3166 2673~2675,2727~2728,2730~2731 PAR:CA:3167 1906,1921~1923,1929~1930 PAR:CA:3168 1858~1861,1878,1880~1881 PAR:CA:3169 536~539,550~551,555 PAR:CA:3170 482,492~495,507~508 |
|
| 1154 |
4woi:DA (3.0Å) |
1539 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS00000ABFE9 | RF00177 RF01959 RF01960 |
Escherichia coli str. K-12 substr. MDS42 1110693 |
26224058 | Assembly2 PAR:DA:1654 1405,1407~1408,1490~1494 PAR:DA:1655 1419~1421,1480 |
|
| 1155 |
4wra:13 (3.05Å) |
1505 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000080E226 | RF00177 RF01959 RF01960 |
Thermus thermophilus HB8 300852 |
26037619 | Assembly1 PAR:13:1745 1405,1408,1490~1491,1493~1495 |
|
| 1156 |
4wra:1G (3.05Å) |
1504 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000080E226 | RF00177 RF01959 RF01960 |
Thermus thermophilus HB8 300852 |
26037619 | Assembly2 PAR:1G:1686 1405~1406,1408,1490~1491,1493~ 1495 |
|
| 1157 |
4wsd:13 (2.95Å) |
1498 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS0000868551 | RF00177 RF01959 RF01960 |
Thermus thermophilus HB8 300852 |
26037619 | Assembly1 PAR:13:1749 1405~1406,1408,1490~1491,1493~ 1495 |
|
| 1158 |
4wsd:1G (2.95Å) |
1498 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS0000868551 | RF00177 RF01959 RF01960 |
Thermus thermophilus HB8 300852 |
26037619 | Assembly2 PAR:1G:1697 1405~1406,1408,1491~1495 |
|
| 1159 |
4wt8:Ab (3.4Å) |
1504 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000080E260 | RF00177 RF01959 RF01960 |
Thermus thermophilus HB8 300852 |
25562208 | Assembly1 PAR:Ab:1601 1388~1389,1391,1469,1471~1472 |
|
| 1160 |
4wt8:Bb (3.4Å) |
1504 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000080E260 | RF00177 RF01959 RF01960 |
Thermus thermophilus HB8 300852 |
25562208 | Assembly2 PAR:Bb:1601 1391,1468~1469,1471~1473 |
|
| 1161 |
4wt8:C1 (3.4Å) |
2807 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000080E136 | RF02540 RF02541 RF00002 |
Thermus thermophilus HB8 300852 |
25562208 | Assembly1 3V6:C1:3001 2085,2449~2450,2611~2613 |
|
| 1162 |
4wt8:C4 (3.4Å) |
77 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000041AF00 | RF00005 | Thermus thermophilus HB8 300852 |
25562208 | Assembly1 3V6:C1:3001 76~77 |
|
| 1163 |
4wt8:D1 (3.4Å) |
2807 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000080E136 | RF02540 RF02541 RF00002 |
Thermus thermophilus HB8 300852 |
25562208 | Assembly2 3V6:D1:3001 2084~2085,2450,2611~2613 |
|
| 1164 |
4www:RA (3.1Å) |
2854 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS00004B0F34 | RF02540 RF02541 |
Escherichia coli str. K-12 substr. MG1655 511145 |
20855725 | Assembly1 EM1:RA:3135 748,750,752,2058~2059,2062,250 3,2505,2609,2611 |
|
| 1165 |
4x4n:B (2.953Å) |
32 | N/A | URS000080E2D0 | N/A | Homo sapiens 9606 |
25640237 |  1124 |
Assembly1 GMP:B:101 15~16 5GP:B:102 14,16~17,19 |
| 1166 |
4x4s:B (3.25Å) |
35 | N/A | URS000080DD43 | N/A | Homo sapiens 9606 |
25640237 |  M044_0.6  1124 |
Assembly1 CTP:A:501 36~37 |
| 1167 |
4x4v:B (2.6Å) |
33 | N/A | URS000080E308 | N/A | Homo sapiens 9606 |
25640237 |  1335  1124  s49  1333 |
Assembly1 TAR:B:501 11,21~22 5GP:B:502 14,16~17,19 5GP:B:503 15~16 |
| 1168 |
4x4v:D (2.6Å) |
26 | N/A | URS000080E308 | N/A | Homo sapiens 9606 |
25640237 | Assembly2 5GP:D:101 16~17 |
|
| 1169 |
4x62:A (3.4492Å) |
1512 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000080DE21 | RF00177 RF01959 RF01960 |
Thermus thermophilus HB8 300852 |
26751643 | Assembly1 PAR:A:1601 1407~1408,1491~1495 PAR:A:1602 569,821 PAR:A:1603 31,47~48,51~52,114,355~356,359 ~360 PAR:A:1604 662,664,666,741~743 PAR:A:1605 610~611,622~626 PAR:A:1606 670~672,716 |
|
| 1170 |
4x64:A (3.35Å) |
1512 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000080DE21 | RF00177 RF01959 RF01960 |
Thermus thermophilus HB8 300852 |
26751643 | Assembly1 PAR:A:1601 1405,1407~1408,1491~1495 PAR:A:1602 568~569,575,821,875 PAR:A:1603 47~48,51~52,114,355~356,359~36 0 PAR:A:1604 662,664,666,741~743 PAR:A:1605 610~611,622~626 PAR:A:1606 669~671,715~716 |
|
| 1171 |
4x65:A (3.345Å) |
1512 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000080DE21 | RF00177 RF01959 RF01960 |
Thermus thermophilus HB8 300852 |
26751643 | Assembly1 PAR:A:1601 1405,1407~1408,1491~1495 PAR:A:1602 569,575,821 PAR:A:1603 47~48,51~52,114,355~356,359~36 0 PAR:A:1604 662,664,740~743 PAR:A:1605 610~611,622~626 PAR:A:1606 670~671,716 |
|
| 1172 |
4x66:A (3.446Å) |
1512 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000080DE21 | RF00177 RF01959 RF01960 |
Thermus thermophilus HB8 300852 |
26751643 | Assembly1 PAR:A:1601 1405,1407~1408,1491,1493~1495 PAR:A:1602 569,575,821 PAR:A:1603 47~48,51~52,114,355~356,359~36 0 PAR:A:1604 662,664,740~743 PAR:A:1605 610~611,622~626 PAR:A:1606 670,672,715~716 |
|
| 1173 |
4xnr:X (2.21Å) |
71 | GO:0009113 (P) purine nucleobase biosy... | URS000080E2A9 | RF00167 | Vibrio vulnificus 672 |
25938715 | Assembly1 ADE:X:101 21~22,47,51~52,74~75 |
|
| 1174 |
4xw0:A (1.81Å) |
10 | N/A | URS00008FED46 | N/A | Homo sapiens 9606 |
26543073 | Assembly1 SO4:A:101 6~7 |
|
| 1175 |
4xw7:A (2.5Å) |
64 | GO:0006730 (P) one-carbon metabolic pr... GO:1900371 (P) regulation of purine nu... |
URS0000868555 | RF01750 | Schaalia odontolytica 1660 |
26144884 | Assembly1 AMZ:A:101 11~12,29 IRI:A:115 7~8 |
|
| 1176 |
4xwf:A (1.8Å) |
64 | GO:0006730 (P) one-carbon metabolic pr... GO:1900371 (P) regulation of purine nu... |
URS0000868555 | RF01750 | Schaalia odontolytica 1660 |
26144884 | Assembly1 AMZ:A:101 11~12,29 IRI:A:105 7~8 IRI:A:106 60~61 IRI:A:107 58~59 IRI:A:108 10,30 |
|
| 1177 |
4y1i:A (2.85Å) |
100 | N/A | URS00008116C1 | N/A | Lactococcus lactis subsp. lactis 1360 |
25794619 | Assembly1 GTP:A:201 3,101 |
|
| 1178 |
4y1i:B (2.85Å) |
100 | N/A | URS00008116C1 | N/A | Lactococcus lactis subsp. lactis 1360 |
25794619 | Assembly1 GTP:A:226 3,101 |
|
| 1179 |
4y1j:A (2.24Å) |
100 | N/A | URS00008116BC | N/A | Lactococcus lactis subsp. lactis 1360 |
25794619 | Assembly1 GTP:A:201 3,101 |
|
| 1180 |
4y1j:B (2.24Å) |
100 | N/A | URS00008116BC | N/A | Lactococcus lactis subsp. lactis 1360 |
25794619 | Assembly1 GTP:B:202 3,101 |
|
| 1181 |
4y1m:A (3.0Å) |
76 | N/A | URS00008116AD | N/A | Escherichia coli 562 |
25794619 | Assembly2 GTP:A:208 2,108 |
|
| 1182 |
4y1m:B (3.0Å) |
107 | GO:0030145 (F) manganese ion binding | URS00008116AD | RF00080 | Escherichia coli 562 |
25794619 | Assembly1 GTP:B:212 2,108 |
|
| 1183 |
4y1n:A (3.0Å) |
249 | GO:0000373 (P) Group II intron splicing | URS0000868528 | RF02001 | Oceanobacillus iheyensis HTE831 221109 |
26502156 | Assembly1 IRI:A:401 8~10,259 IRI:A:402 70,113~114 IRI:A:403 25,27~29 IRI:A:404 12,14~16,255~256 IRI:A:406 36~37 IRI:A:407 39,243~244 IRI:A:408 242~244 IRI:A:409 40~41 IRI:A:410 119,173 IRI:A:411 264~266 IRI:A:412 31~32 IRI:A:413 164,211~213 |
|
| 1184 |
4y1n:B (3.0Å) |
259 | GO:0000373 (P) Group II intron splicing | URS0000868528 | RF02001 | Oceanobacillus iheyensis HTE831 221109 |
26502156 | Assembly2 IRI:B:401 32,34 IRI:B:402 12,14~15,255 IRI:B:403 8~10,259 IRI:B:404 38~39,243~244 IRI:B:405 242~244 IRI:B:407 80,100~101 IRI:B:408 212~213 IRI:B:409 159~160 IRI:B:410 264~266 IRI:B:411 31~32 IRI:B:412 52~53,169~170 IRI:B:413 64,120 IRI:B:414 119,239~240 IRI:B:415 142,144~146 |
|
| 1185 |
4y4o:1A (2.3Å) |
2872 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS00001698E8 | RF02540 RF02541 RF00002 |
Thermus thermophilus HB8 300852 |
25775268 | Assembly1 MPD:1A:4028 449~451 |
|
| 1186 |
4y4o:1B (2.3Å) |
120 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000080DEE8 | RF00001 | Thermus thermophilus HB8 300852 |
25775268 | Assembly1 ARG:1B:230 80~82,93~94 |
|
| 1187 |
4y4o:1a (2.3Å) |
1500 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000080DFED | RF00177 RF01959 RF01960 |
Thermus thermophilus HB8 300852 |
25775268 | Assembly1 MPD:1a:1880 345~347 |
|
| 1188 |
4y4o:2A (2.3Å) |
2867 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS00001698E8 | RF02540 RF02541 RF00002 |
Thermus thermophilus HB8 300852 |
25775268 | Assembly2 MPD:2A:3747 1772,1976,1978 MPD:2A:3748 2397,2413~2414 MPD:2B:3020 915~916 |
|
| 1189 |
4yaz:A (2.0Å) |
84 | GO:0035438 (F) cyclic-di-GMP binding | URS00008116B1 | RF01051 | Geobacter 28231 |
25818298 | Assembly2 4BW:A:103 8,11~12,14,40~41,75~76 |
|
| 1190 |
4yaz:R (2.0Å) |
84 | GO:0035438 (F) cyclic-di-GMP binding | URS00008116B1 | RF01051 | Geobacter 28231 |
25818298 | Assembly1 4BW:R:106 8,11~12,14,40~41,75~76 |
|
| 1191 |
4yb0:A (2.121Å) |
83 | GO:0035438 (F) cyclic-di-GMP binding | URS00008116A8 | RF01051 | Geobacter 28231 |
25818298 | Assembly2 GDP:A:101 2,84 C2E:A:102 8,11~12,14,40~41,75~76 |
|
| 1192 |
4yb0:R (2.121Å) |
83 | GO:0035438 (F) cyclic-di-GMP binding | URS00008116A8 | RF01051 | Geobacter 28231 |
25818298 | Assembly1 GDP:R:101 2,84 C2E:R:102 8,11~12,14,40~41,75~76 |
|
| 1193 |
4yb1:R (2.081Å) |
91 | GO:0035438 (F) cyclic-di-GMP binding | URS00008116BA | RF03167 RF01051 |
Vibrio cholerae 666 |
25818298 | Assembly1 4BW:R:102 14,16~17,20~21,46~47,93~94 |
|
| 1194 |
4ybb:AA (2.1Å) |
1534 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000081169C | RF00177 RF01959 RF01960 |
Escherichia coli K-12 83333 |
25775265 | Assembly1 PG4:AA:1670 774~775,806 MPD:AA:1671 1457~1458 MPD:AA:1676 293,307~308 PUT:AA:1673 453,479~480 |
|
| 1195 |
4ybb:BA (2.1Å) |
1533 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000081169C | RF00177 RF01959 RF01960 |
Escherichia coli K-12 83333 |
25775265 | Assembly2 PG4:BA:1642 774,806 |
|
| 1196 |
4ybb:DA (2.1Å) |
2897 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS000080E25C | RF02540 RF02541 |
Escherichia coli K-12 83333 |
25775265 | Assembly1 SPD:DA:3183 855~857 SPD:DA:3187 860~861,910~911,913,2265 SPD:DA:3207 1769~1770,1959,1975 SPD:DA:3225 677,792~793,2440~2442 1PE:DA:3185 22~23,519 1PE:DA:3203 948~949,961 PGE:DA:3186 998~999,1158 PGE:DA:3215 1424,1575 PGE:DA:3226 1693,1831,1975~1976 PUT:DA:3189 504~505,1235~1236 PUT:DA:3195 2628~2629,2781 PUT:DA:3206 1904,1968,2593 PUT:DA:3213 1698,1979 PUT:DA:3214 1132~1133,1136~1137 PUT:DA:3220 698,1554~1555 PUT:DA:3223 787~790,794 PUT:DA:3224 825,2259,2428 MPD:DA:3190 602,655 MPD:DA:3192 1202,1244 MPD:DA:3205 61~63,92~93 MPD:DA:3208 2751~2752 MPD:DA:3211 2451~2452,2506 ACY:DA:3191 59~60,74 PG4:DA:3193 2256~2258,2275 PG4:DA:3217 1349,1383,1406 EDO:DA:3194 1782,2609 EDO:DA:3197 189,205 EDO:DA:3198 2821,2826~2827 EDO:DR:203 998~999 PEG:DA:3200 2008~2009 PEG:DA:3201 978~979,986,1002 PEG:DA:3227 747,2015 PEG:DA:3228 2058~2059 GUN:DA:3212 2814,2831 |
|
| 1197 |
4ybb:DB (2.1Å) |
120 | GO:0003735 (F) structural constituent ... GO:0005840 (C) ribosome |
URS0000049E57 | RF00001 | Escherichia coli K-12 83333 |
25775265 | Assembly1 EDO:DB:212 84~85,93 |
|
| 1198 |
4yco:D (2.1Å) |
74 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000080E264 | RF00005 | Escherichia coli K-12 83333 |
25902496 | Assembly1 FMN:A:401 16~17 |
|
| 1199 |
4yco:E (2.1Å) |
67 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000080E264 | RF00005 | Escherichia coli K-12 83333 |
25902496 | Assembly2 FMN:B:402 16~17 |
|
| 1200 |
4yco:F (2.1Å) |
71 | GO:0030533 (F) triplet codon-amino aci... GO:0006412 (P) translation |
URS000080E264 | RF00005 | Escherichia coli K-12 83333 |
25902496 | Assembly3 FMN:C:401 16~17 |

Download all results in tab-seperated text for
3166 RNAs.
Click the entry in the '#' column to view structure details of that RNA chain.
Hover over PDB to view the title of the structure.
Hover over Rfam to view names of Rfam families.
Hover over PubMed to view title of the PubMed publications.
Hover over Ligand to view the name of the ligand and the ligand-binding nucleotides on the RNA.
<<
<
2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22
>
>>
<<
<
2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22
>
>>
Reference:
